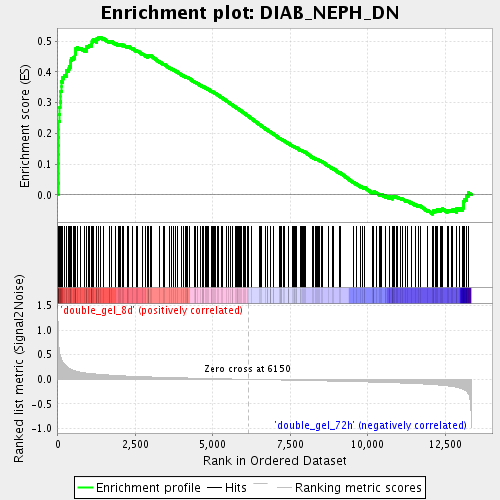
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | DIAB_NEPH_DN |
| Enrichment Score (ES) | 0.5125446 |
| Normalized Enrichment Score (NES) | 2.0672379 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0028107276 |
| FWER p-Value | 0.065 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IGFBP7 | IGFBP7 Entrez, Source | insulin-like growth factor binding protein 7 | 5 | 0.972 | 0.0386 | Yes |
| 2 | ANXA1 | ANXA1 Entrez, Source | annexin A1 | 10 | 0.815 | 0.0709 | Yes |
| 3 | SERPINE1 | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 12 | 0.803 | 0.1030 | Yes |
| 4 | COL3A1 | COL3A1 Entrez, Source | collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | 15 | 0.737 | 0.1324 | Yes |
| 5 | ARMCX2 | ARMCX2 Entrez, Source | armadillo repeat containing, X-linked 2 | 19 | 0.707 | 0.1605 | Yes |
| 6 | LPL | LPL Entrez, Source | lipoprotein lipase | 24 | 0.680 | 0.1875 | Yes |
| 7 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 32 | 0.660 | 0.2134 | Yes |
| 8 | ADM | ADM Entrez, Source | adrenomedullin | 34 | 0.649 | 0.2393 | Yes |
| 9 | GPRC5A | GPRC5A Entrez, Source | G protein-coupled receptor, family C, group 5, member A | 49 | 0.587 | 0.2618 | Yes |
| 10 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 59 | 0.557 | 0.2834 | Yes |
| 11 | PRKAR2B | PRKAR2B Entrez, Source | protein kinase, cAMP-dependent, regulatory, type II, beta | 69 | 0.494 | 0.3025 | Yes |
| 12 | F2R | F2R Entrez, Source | coagulation factor II (thrombin) receptor | 89 | 0.456 | 0.3193 | Yes |
| 13 | BMP2 | BMP2 Entrez, Source | bone morphogenetic protein 2 | 91 | 0.451 | 0.3373 | Yes |
| 14 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 114 | 0.408 | 0.3520 | Yes |
| 15 | CXCL2 | CXCL2 Entrez, Source | chemokine (C-X-C motif) ligand 2 | 118 | 0.402 | 0.3679 | Yes |
| 16 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 145 | 0.367 | 0.3806 | Yes |
| 17 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 196 | 0.324 | 0.3897 | Yes |
| 18 | TSC22D3 | TSC22D3 Entrez, Source | TSC22 domain family, member 3 | 276 | 0.263 | 0.3942 | Yes |
| 19 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 280 | 0.261 | 0.4045 | Yes |
| 20 | ENPEP | ENPEP Entrez, Source | glutamyl aminopeptidase (aminopeptidase A) | 338 | 0.234 | 0.4095 | Yes |
| 21 | PTGER4 | PTGER4 Entrez, Source | prostaglandin E receptor 4 (subtype EP4) | 361 | 0.225 | 0.4168 | Yes |
| 22 | GRK5 | GRK5 Entrez, Source | G protein-coupled receptor kinase 5 | 401 | 0.209 | 0.4222 | Yes |
| 23 | CDS1 | CDS1 Entrez, Source | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 | 409 | 0.207 | 0.4300 | Yes |
| 24 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 418 | 0.205 | 0.4376 | Yes |
| 25 | DNAJB9 | DNAJB9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 9 | 437 | 0.199 | 0.4442 | Yes |
| 26 | DPYSL3 | DPYSL3 Entrez, Source | dihydropyrimidinase-like 3 | 499 | 0.182 | 0.4468 | Yes |
| 27 | ADORA1 | ADORA1 Entrez, Source | adenosine A1 receptor | 547 | 0.173 | 0.4502 | Yes |
| 28 | TMEM123 | TMEM123 Entrez, Source | transmembrane protein 123 | 551 | 0.172 | 0.4568 | Yes |
| 29 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 559 | 0.171 | 0.4631 | Yes |
| 30 | SCRN1 | SCRN1 Entrez, Source | secernin 1 | 568 | 0.170 | 0.4694 | Yes |
| 31 | TGFBR3 | TGFBR3 Entrez, Source | transforming growth factor, beta receptor III (betaglycan, 300kDa) | 577 | 0.168 | 0.4755 | Yes |
| 32 | PDGFA | PDGFA Entrez, Source | platelet-derived growth factor alpha polypeptide | 619 | 0.162 | 0.4789 | Yes |
| 33 | COLQ | COLQ Entrez, Source | collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | 718 | 0.149 | 0.4773 | Yes |
| 34 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 858 | 0.133 | 0.4721 | Yes |
| 35 | CALD1 | CALD1 Entrez, Source | caldesmon 1 | 920 | 0.127 | 0.4725 | Yes |
| 36 | FGF9 | FGF9 Entrez, Source | fibroblast growth factor 9 (glia-activating factor) | 921 | 0.127 | 0.4776 | Yes |
| 37 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 930 | 0.126 | 0.4821 | Yes |
| 38 | CLIC5 | CLIC5 Entrez, Source | chloride intracellular channel 5 | 976 | 0.123 | 0.4836 | Yes |
| 39 | RGS10 | RGS10 Entrez, Source | regulator of G-protein signalling 10 | 1009 | 0.121 | 0.4860 | Yes |
| 40 | PLSCR1 | PLSCR1 Entrez, Source | phospholipid scramblase 1 | 1073 | 0.116 | 0.4858 | Yes |
| 41 | PTPRD | PTPRD Entrez, Source | protein tyrosine phosphatase, receptor type, D | 1088 | 0.116 | 0.4894 | Yes |
| 42 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 1098 | 0.115 | 0.4933 | Yes |
| 43 | PPFIA4 | PPFIA4 Entrez, Source | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | 1099 | 0.115 | 0.4979 | Yes |
| 44 | PLCG2 | PLCG2 Entrez, Source | phospholipase C, gamma 2 (phosphatidylinositol-specific) | 1116 | 0.114 | 0.5012 | Yes |
| 45 | LOXL1 | LOXL1 Entrez, Source | lysyl oxidase-like 1 | 1137 | 0.113 | 0.5042 | Yes |
| 46 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 1250 | 0.107 | 0.5000 | Yes |
| 47 | TSPAN3 | TSPAN3 Entrez, Source | tetraspanin 3 | 1254 | 0.107 | 0.5041 | Yes |
| 48 | CAST | CAST Entrez, Source | calpastatin | 1258 | 0.107 | 0.5081 | Yes |
| 49 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 1297 | 0.104 | 0.5094 | Yes |
| 50 | MYO1B | MYO1B Entrez, Source | myosin IB | 1311 | 0.104 | 0.5125 | Yes |
| 51 | COL4A4 | COL4A4 Entrez, Source | collagen, type IV, alpha 4 | 1373 | 0.100 | 0.5119 | No |
| 52 | SULF1 | SULF1 Entrez, Source | sulfatase 1 | 1470 | 0.095 | 0.5084 | No |
| 53 | APOD | APOD Entrez, Source | apolipoprotein D | 1653 | 0.087 | 0.4980 | No |
| 54 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 1672 | 0.086 | 0.5000 | No |
| 55 | CAPN2 | CAPN2 Entrez, Source | calpain 2, (m/II) large subunit | 1726 | 0.084 | 0.4993 | No |
| 56 | SERINC5 | SERINC5 Entrez, Source | serine incorporator 5 | 1858 | 0.078 | 0.4925 | No |
| 57 | MYO1E | MYO1E Entrez, Source | myosin IE | 1942 | 0.075 | 0.4891 | No |
| 58 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 1975 | 0.074 | 0.4896 | No |
| 59 | PEX14 | PEX14 Entrez, Source | peroxisomal biogenesis factor 14 | 2007 | 0.073 | 0.4902 | No |
| 60 | TMED5 | TMED5 Entrez, Source | transmembrane emp24 protein transport domain containing 5 | 2090 | 0.070 | 0.4867 | No |
| 61 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 2118 | 0.069 | 0.4875 | No |
| 62 | CXADR | CXADR Entrez, Source | coxsackie virus and adenovirus receptor | 2231 | 0.066 | 0.4816 | No |
| 63 | ATXN1 | ATXN1 Entrez, Source | ataxin 1 | 2254 | 0.066 | 0.4825 | No |
| 64 | ING1 | ING1 Entrez, Source | inhibitor of growth family, member 1 | 2289 | 0.065 | 0.4825 | No |
| 65 | PAWR | PAWR Entrez, Source | PRKC, apoptosis, WT1, regulator | 2421 | 0.061 | 0.4750 | No |
| 66 | DDN | DDN Entrez, Source | dendrin | 2535 | 0.059 | 0.4687 | No |
| 67 | SLIT1 | SLIT1 Entrez, Source | slit homolog 1 (Drosophila) | 2578 | 0.058 | 0.4678 | No |
| 68 | PSMA6 | PSMA6 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 6 | 2734 | 0.054 | 0.4582 | No |
| 69 | SYNPO | SYNPO Entrez, Source | synaptopodin | 2831 | 0.052 | 0.4529 | No |
| 70 | PRKAR1A | PRKAR1A Entrez, Source | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 2902 | 0.051 | 0.4496 | No |
| 71 | ARL1 | ARL1 Entrez, Source | ADP-ribosylation factor-like 1 | 2908 | 0.051 | 0.4513 | No |
| 72 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 2919 | 0.050 | 0.4525 | No |
| 73 | NPTN | NPTN Entrez, Source | neuroplastin | 2925 | 0.050 | 0.4542 | No |
| 74 | BCAM | BCAM Entrez, Source | basal cell adhesion molecule (Lutheran blood group) | 2988 | 0.049 | 0.4514 | No |
| 75 | PLEKHC1 | PLEKHC1 Entrez, Source | pleckstrin homology domain containing, family C (with FERM domain) member 1 | 3006 | 0.048 | 0.4520 | No |
| 76 | SKP1A | SKP1A Entrez, Source | S-phase kinase-associated protein 1A (p19A) | 3287 | 0.043 | 0.4324 | No |
| 77 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 3405 | 0.040 | 0.4251 | No |
| 78 | PTHR1 | PTHR1 Entrez, Source | parathyroid hormone receptor 1 | 3437 | 0.040 | 0.4243 | No |
| 79 | GTF2A2 | GTF2A2 Entrez, Source | general transcription factor IIA, 2, 12kDa | 3615 | 0.037 | 0.4123 | No |
| 80 | LITAF | LITAF Entrez, Source | lipopolysaccharide-induced TNF factor | 3654 | 0.036 | 0.4108 | No |
| 81 | ITGB5 | ITGB5 Entrez, Source | integrin, beta 5 | 3714 | 0.035 | 0.4077 | No |
| 82 | HTRA1 | HTRA1 Entrez, Source | HtrA serine peptidase 1 | 3790 | 0.034 | 0.4033 | No |
| 83 | CTBP1 | CTBP1 Entrez, Source | C-terminal binding protein 1 | 3848 | 0.033 | 0.4003 | No |
| 84 | GSTA4 | GSTA4 Entrez, Source | glutathione S-transferase A4 | 3997 | 0.030 | 0.3902 | No |
| 85 | AIF1 | AIF1 Entrez, Source | allograft inflammatory factor 1 | 4047 | 0.030 | 0.3876 | No |
| 86 | ABI1 | ABI1 Entrez, Source | abl-interactor 1 | 4112 | 0.029 | 0.3839 | No |
| 87 | OAS1 | OAS1 Entrez, Source | 2',5'-oligoadenylate synthetase 1, 40/46kDa | 4150 | 0.028 | 0.3822 | No |
| 88 | PPP2CB | PPP2CB Entrez, Source | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | 4166 | 0.028 | 0.3822 | No |
| 89 | TNNT2 | TNNT2 Entrez, Source | troponin T type 2 (cardiac) | 4182 | 0.027 | 0.3821 | No |
| 90 | PHYHIP | PHYHIP Entrez, Source | phytanoyl-CoA 2-hydroxylase interacting protein | 4233 | 0.026 | 0.3794 | No |
| 91 | RARRES2 | RARRES2 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 2 | 4407 | 0.024 | 0.3671 | No |
| 92 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 4430 | 0.023 | 0.3664 | No |
| 93 | STX7 | STX7 Entrez, Source | syntaxin 7 | 4452 | 0.023 | 0.3657 | No |
| 94 | PTPRO | PTPRO Entrez, Source | protein tyrosine phosphatase, receptor type, O | 4511 | 0.022 | 0.3621 | No |
| 95 | CD164 | CD164 Entrez, Source | CD164 molecule, sialomucin | 4616 | 0.021 | 0.3550 | No |
| 96 | HSP90AA1 | HSP90AA1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class A member 1 | 4655 | 0.020 | 0.3530 | No |
| 97 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 4681 | 0.020 | 0.3518 | No |
| 98 | CALM2 | CALM2 Entrez, Source | calmodulin 2 (phosphorylase kinase, delta) | 4699 | 0.020 | 0.3513 | No |
| 99 | TMEM23 | TMEM23 Entrez, Source | transmembrane protein 23 | 4710 | 0.019 | 0.3513 | No |
| 100 | TOMM20 | TOMM20 Entrez, Source | translocase of outer mitochondrial membrane 20 homolog (yeast) | 4757 | 0.019 | 0.3486 | No |
| 101 | DNM1L | DNM1L Entrez, Source | dynamin 1-like | 4783 | 0.018 | 0.3474 | No |
| 102 | PDIA3 | PDIA3 Entrez, Source | protein disulfide isomerase family A, member 3 | 4843 | 0.018 | 0.3436 | No |
| 103 | CFLAR | CFLAR Entrez, Source | CASP8 and FADD-like apoptosis regulator | 4863 | 0.017 | 0.3429 | No |
| 104 | DPP6 | DPP6 Entrez, Source | dipeptidyl-peptidase 6 | 4957 | 0.016 | 0.3364 | No |
| 105 | ACBD3 | ACBD3 Entrez, Source | acyl-Coenzyme A binding domain containing 3 | 4964 | 0.016 | 0.3366 | No |
| 106 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 4986 | 0.015 | 0.3356 | No |
| 107 | DSTN | DSTN Entrez, Source | destrin (actin depolymerizing factor) | 5007 | 0.015 | 0.3347 | No |
| 108 | CD47 | CD47 Entrez, Source | CD47 molecule | 5066 | 0.014 | 0.3308 | No |
| 109 | HMGN3 | HMGN3 Entrez, Source | high mobility group nucleosomal binding domain 3 | 5092 | 0.014 | 0.3295 | No |
| 110 | MAPK10 | MAPK10 Entrez, Source | mitogen-activated protein kinase 10 | 5148 | 0.013 | 0.3258 | No |
| 111 | VPS26A | VPS26A Entrez, Source | vacuolar protein sorting 26 homolog A (yeast) | 5185 | 0.013 | 0.3236 | No |
| 112 | CEBPD | CEBPD Entrez, Source | CCAAT/enhancer binding protein (C/EBP), delta | 5268 | 0.012 | 0.3178 | No |
| 113 | OPTN | OPTN Entrez, Source | optineurin | 5299 | 0.011 | 0.3159 | No |
| 114 | RHOBTB2 | RHOBTB2 Entrez, Source | Rho-related BTB domain containing 2 | 5450 | 0.009 | 0.3049 | No |
| 115 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 5501 | 0.008 | 0.3014 | No |
| 116 | CACNB2 | CACNB2 Entrez, Source | calcium channel, voltage-dependent, beta 2 subunit | 5571 | 0.007 | 0.2964 | No |
| 117 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 5627 | 0.007 | 0.2925 | No |
| 118 | AXL | AXL Entrez, Source | AXL receptor tyrosine kinase | 5649 | 0.006 | 0.2911 | No |
| 119 | PFN2 | PFN2 Entrez, Source | profilin 2 | 5738 | 0.005 | 0.2846 | No |
| 120 | NBN | NBN Entrez, Source | nibrin | 5769 | 0.005 | 0.2825 | No |
| 121 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 5788 | 0.005 | 0.2813 | No |
| 122 | TBCA | TBCA Entrez, Source | tubulin-specific chaperone a | 5793 | 0.005 | 0.2812 | No |
| 123 | HNRPDL | HNRPDL Entrez, Source | heterogeneous nuclear ribonucleoprotein D-like | 5830 | 0.004 | 0.2786 | No |
| 124 | PLS3 | PLS3 Entrez, Source | plastin 3 (T isoform) | 5853 | 0.004 | 0.2771 | No |
| 125 | GAB2 | GAB2 Entrez, Source | GRB2-associated binding protein 2 | 5899 | 0.003 | 0.2738 | No |
| 126 | RHEB | RHEB Entrez, Source | Ras homolog enriched in brain | 5918 | 0.003 | 0.2726 | No |
| 127 | CMAH | CMAH Entrez, Source | cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) | 5991 | 0.002 | 0.2672 | No |
| 128 | ATXN10 | ATXN10 Entrez, Source | ataxin 10 | 6035 | 0.002 | 0.2639 | No |
| 129 | PRKCI | PRKCI Entrez, Source | protein kinase C, iota | 6041 | 0.002 | 0.2636 | No |
| 130 | NDN | NDN Entrez, Source | necdin homolog (mouse) | 6108 | 0.001 | 0.2586 | No |
| 131 | TAX1BP1 | TAX1BP1 Entrez, Source | Tax1 (human T-cell leukemia virus type I) binding protein 1 | 6136 | 0.000 | 0.2566 | No |
| 132 | ATP6AP2 | ATP6AP2 Entrez, Source | ATPase, H+ transporting, lysosomal accessory protein 2 | 6241 | -0.001 | 0.2487 | No |
| 133 | CANX | CANX Entrez, Source | calnexin | 6492 | -0.004 | 0.2298 | No |
| 134 | MGAT5 | MGAT5 Entrez, Source | mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase | 6546 | -0.005 | 0.2259 | No |
| 135 | HSPA5 | HSPA5 Entrez, Source | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 6557 | -0.005 | 0.2254 | No |
| 136 | DAG1 | DAG1 Entrez, Source | dystroglycan 1 (dystrophin-associated glycoprotein 1) | 6712 | -0.007 | 0.2139 | No |
| 137 | WT1 | WT1 Entrez, Source | Wilms tumor 1 | 6748 | -0.008 | 0.2115 | No |
| 138 | TMED10 | TMED10 Entrez, Source | transmembrane emp24-like trafficking protein 10 (yeast) | 6770 | -0.008 | 0.2102 | No |
| 139 | TCF21 | TCF21 Entrez, Source | transcription factor 21 | 6846 | -0.009 | 0.2049 | No |
| 140 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 6866 | -0.009 | 0.2038 | No |
| 141 | NAB1 | NAB1 Entrez, Source | NGFI-A binding protein 1 (EGR1 binding protein 1) | 6874 | -0.009 | 0.2036 | No |
| 142 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 6962 | -0.010 | 0.1974 | No |
| 143 | SMARCA2 | SMARCA2 Entrez, Source | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 6971 | -0.010 | 0.1972 | No |
| 144 | SACM1L | SACM1L Entrez, Source | SAC1 suppressor of actin mutations 1-like (yeast) | 7162 | -0.013 | 0.1832 | No |
| 145 | NPHS1 | NPHS1 Entrez, Source | nephrosis 1, congenital, Finnish type (nephrin) | 7169 | -0.013 | 0.1832 | No |
| 146 | CASP1 | CASP1 Entrez, Source | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | 7177 | -0.013 | 0.1832 | No |
| 147 | HMGN1 | HMGN1 Entrez, Source | high-mobility group nucleosome binding domain 1 | 7215 | -0.013 | 0.1809 | No |
| 148 | EPS15 | EPS15 Entrez, Source | epidermal growth factor receptor pathway substrate 15 | 7217 | -0.013 | 0.1814 | No |
| 149 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 7272 | -0.014 | 0.1778 | No |
| 150 | NTRK2 | NTRK2 Entrez, Source | neurotrophic tyrosine kinase, receptor, type 2 | 7314 | -0.014 | 0.1753 | No |
| 151 | SEPT2 | SEPT2 Entrez, Source | septin 2 | 7427 | -0.016 | 0.1673 | No |
| 152 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 7435 | -0.016 | 0.1674 | No |
| 153 | CREB3L2 | CREB3L2 Entrez, Source | cAMP responsive element binding protein 3-like 2 | 7447 | -0.016 | 0.1672 | No |
| 154 | UBE2L6 | UBE2L6 Entrez, Source | ubiquitin-conjugating enzyme E2L 6 | 7569 | -0.018 | 0.1587 | No |
| 155 | PSMA2 | PSMA2 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 2 | 7610 | -0.018 | 0.1564 | No |
| 156 | MME | MME Entrez, Source | membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase) | 7620 | -0.018 | 0.1565 | No |
| 157 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 7649 | -0.019 | 0.1551 | No |
| 158 | DNAJA1 | DNAJA1 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 1 | 7683 | -0.019 | 0.1533 | No |
| 159 | GYG1 | GYG1 Entrez, Source | glycogenin 1 | 7708 | -0.019 | 0.1523 | No |
| 160 | CAND2 | CAND2 Entrez, Source | cullin-associated and neddylation-dissociated 2 (putative) | 7711 | -0.020 | 0.1529 | No |
| 161 | NAPG | NAPG Entrez, Source | N-ethylmaleimide-sensitive factor attachment protein, gamma | 7833 | -0.021 | 0.1445 | No |
| 162 | GMFB | GMFB Entrez, Source | glia maturation factor, beta | 7835 | -0.021 | 0.1453 | No |
| 163 | KLK6 | KLK6 Entrez, Source | kallikrein 6 (neurosin, zyme) | 7846 | -0.021 | 0.1454 | No |
| 164 | ADD3 | ADD3 Entrez, Source | adducin 3 (gamma) | 7901 | -0.022 | 0.1421 | No |
| 165 | SUB1 | SUB1 Entrez, Source | SUB1 homolog (S. cerevisiae) | 7922 | -0.022 | 0.1415 | No |
| 166 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 7961 | -0.023 | 0.1395 | No |
| 167 | AQP3 | AQP3 Entrez, Source | aquaporin 3 (Gill blood group) | 7975 | -0.023 | 0.1394 | No |
| 168 | GRIK2 | GRIK2 Entrez, Source | glutamate receptor, ionotropic, kainate 2 | 8215 | -0.027 | 0.1223 | No |
| 169 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8261 | -0.027 | 0.1199 | No |
| 170 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 8322 | -0.028 | 0.1165 | No |
| 171 | YWHAZ | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 8338 | -0.028 | 0.1165 | No |
| 172 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 8373 | -0.029 | 0.1150 | No |
| 173 | BIRC3 | BIRC3 Entrez, Source | baculoviral IAP repeat-containing 3 | 8408 | -0.029 | 0.1136 | No |
| 174 | IRS1 | IRS1 Entrez, Source | insulin receptor substrate 1 | 8456 | -0.030 | 0.1113 | No |
| 175 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 8496 | -0.031 | 0.1095 | No |
| 176 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 8535 | -0.031 | 0.1079 | No |
| 177 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 8731 | -0.034 | 0.0943 | No |
| 178 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 8852 | -0.035 | 0.0866 | No |
| 179 | NEDD9 | NEDD9 Entrez, Source | neural precursor cell expressed, developmentally down-regulated 9 | 8900 | -0.036 | 0.0845 | No |
| 180 | CYP51A1 | CYP51A1 Entrez, Source | cytochrome P450, family 51, subfamily A, polypeptide 1 | 9103 | -0.039 | 0.0706 | No |
| 181 | HNRPC | HNRPC Entrez, Source | heterogeneous nuclear ribonucleoprotein C (C1/C2) | 9106 | -0.039 | 0.0720 | No |
| 182 | PDE4B | PDE4B Entrez, Source | phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) | 9537 | -0.046 | 0.0410 | No |
| 183 | ARF6 | ARF6 Entrez, Source | ADP-ribosylation factor 6 | 9627 | -0.047 | 0.0362 | No |
| 184 | PLXNB2 | PLXNB2 Entrez, Source | plexin B2 | 9773 | -0.050 | 0.0271 | No |
| 185 | BIRC2 | BIRC2 Entrez, Source | baculoviral IAP repeat-containing 2 | 9826 | -0.051 | 0.0251 | No |
| 186 | PTPN11 | PTPN11 Entrez, Source | protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) | 9881 | -0.052 | 0.0231 | No |
| 187 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 9899 | -0.052 | 0.0239 | No |
| 188 | PTPN7 | PTPN7 Entrez, Source | protein tyrosine phosphatase, non-receptor type 7 | 10142 | -0.056 | 0.0076 | No |
| 189 | PCMT1 | PCMT1 Entrez, Source | protein-L-isoaspartate (D-aspartate) O-methyltransferase | 10182 | -0.056 | 0.0069 | No |
| 190 | MAGI2 | MAGI2 Entrez, Source | membrane associated guanylate kinase, WW and PDZ domain containing 2 | 10184 | -0.056 | 0.0091 | No |
| 191 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 10199 | -0.057 | 0.0103 | No |
| 192 | PBEF1 | PBEF1 Entrez, Source | pre-B-cell colony enhancing factor 1 | 10279 | -0.058 | 0.0066 | No |
| 193 | ARHGAP5 | ARHGAP5 Entrez, Source | Rho GTPase activating protein 5 | 10392 | -0.060 | 0.0005 | No |
| 194 | CSPG2 | CSPG2 Entrez, Source | chondroitin sulfate proteoglycan 2 (versican) | 10417 | -0.061 | 0.0011 | No |
| 195 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 10450 | -0.061 | 0.0011 | No |
| 196 | BMP7 | BMP7 Entrez, Source | bone morphogenetic protein 7 (osteogenic protein 1) | 10568 | -0.064 | -0.0053 | No |
| 197 | GLUL | GLUL Entrez, Source | glutamate-ammonia ligase (glutamine synthetase) | 10574 | -0.064 | -0.0031 | No |
| 198 | VEGF | VEGF Entrez, Source | vascular endothelial growth factor | 10686 | -0.066 | -0.0090 | No |
| 199 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 10688 | -0.066 | -0.0064 | No |
| 200 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 10785 | -0.068 | -0.0110 | No |
| 201 | LANCL1 | LANCL1 Entrez, Source | LanC lantibiotic synthetase component C-like 1 (bacterial) | 10788 | -0.068 | -0.0084 | No |
| 202 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 10803 | -0.068 | -0.0067 | No |
| 203 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 10816 | -0.069 | -0.0049 | No |
| 204 | EXOSC7 | EXOSC7 Entrez, Source | exosome component 7 | 10861 | -0.070 | -0.0055 | No |
| 205 | RAI14 | RAI14 Entrez, Source | retinoic acid induced 14 | 10917 | -0.071 | -0.0068 | No |
| 206 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 10964 | -0.073 | -0.0074 | No |
| 207 | RDX | RDX Entrez, Source | radixin | 11042 | -0.074 | -0.0103 | No |
| 208 | IGFBP2 | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 11121 | -0.076 | -0.0132 | No |
| 209 | FNBP1 | FNBP1 Entrez, Source | formin binding protein 1 | 11225 | -0.079 | -0.0179 | No |
| 210 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 11295 | -0.081 | -0.0199 | No |
| 211 | SDC2 | SDC2 Entrez, Source | syndecan 2 (heparan sulfate proteoglycan 1, cell surface-associated, fibroglycan) | 11419 | -0.084 | -0.0259 | No |
| 212 | NFASC | NFASC Entrez, Source | neurofascin homolog (chicken) | 11534 | -0.087 | -0.0311 | No |
| 213 | RAB21 | RAB21 Entrez, Source | RAB21, member RAS oncogene family | 11632 | -0.090 | -0.0349 | No |
| 214 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 11694 | -0.092 | -0.0359 | No |
| 215 | NMI | NMI Entrez, Source | N-myc (and STAT) interactor | 11925 | -0.100 | -0.0494 | No |
| 216 | SLK | SLK Entrez, Source | STE20-like kinase (yeast) | 12099 | -0.107 | -0.0584 | No |
| 217 | TAPBP | TAPBP Entrez, Source | TAP binding protein (tapasin) | 12109 | -0.108 | -0.0547 | No |
| 218 | PODXL | PODXL Entrez, Source | podocalyxin-like | 12132 | -0.109 | -0.0521 | No |
| 219 | AGRIN | AGRIN Entrez, Source | agrin | 12183 | -0.111 | -0.0514 | No |
| 220 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 12232 | -0.113 | -0.0506 | No |
| 221 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 12235 | -0.113 | -0.0462 | No |
| 222 | BST2 | BST2 Entrez, Source | bone marrow stromal cell antigen 2 | 12353 | -0.119 | -0.0504 | No |
| 223 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 12371 | -0.119 | -0.0469 | No |
| 224 | GPNMB | GPNMB Entrez, Source | glycoprotein (transmembrane) nmb | 12406 | -0.122 | -0.0446 | No |
| 225 | ANKRD15 | ANKRD15 Entrez, Source | ankyrin repeat domain 15 | 12561 | -0.132 | -0.0511 | No |
| 226 | MECP2 | MECP2 Entrez, Source | methyl CpG binding protein 2 (Rett syndrome) | 12612 | -0.135 | -0.0495 | No |
| 227 | EXOC7 | EXOC7 Entrez, Source | exocyst complex component 7 | 12695 | -0.141 | -0.0501 | No |
| 228 | GHR | GHR Entrez, Source | growth hormone receptor | 12744 | -0.146 | -0.0479 | No |
| 229 | KLK7 | KLK7 Entrez, Source | kallikrein 7 (chymotryptic, stratum corneum) | 12860 | -0.160 | -0.0503 | No |
| 230 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 12862 | -0.160 | -0.0439 | No |
| 231 | AKAP11 | AKAP11 Entrez, Source | A kinase (PRKA) anchor protein 11 | 12961 | -0.175 | -0.0444 | No |
| 232 | TRIM23 | TRIM23 Entrez, Source | tripartite motif-containing 23 | 13053 | -0.198 | -0.0434 | No |
| 233 | PSMB9 | PSMB9 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) | 13079 | -0.204 | -0.0372 | No |
| 234 | TSPYL4 | TSPYL4 Entrez, Source | TSPY-like 4 | 13090 | -0.207 | -0.0296 | No |
| 235 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13096 | -0.208 | -0.0217 | No |
| 236 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13114 | -0.214 | -0.0144 | No |
| 237 | GULP1 | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 13181 | -0.242 | -0.0097 | No |
| 238 | FGFR2 | FGFR2 Entrez, Source | ffer syndrome, Jackson-Weiss syndrome) | 13200 | -0.255 | -0.0009 | No |
| 239 | OTUD4 | OTUD4 Entrez, Source | OTU domain containing 4 | 13243 | -0.291 | 0.0076 | No |