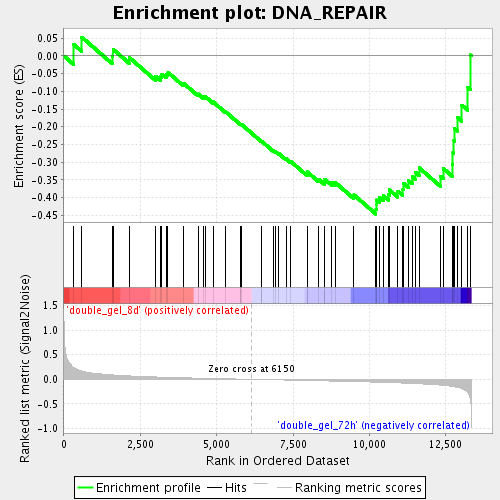
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | DNA_REPAIR |
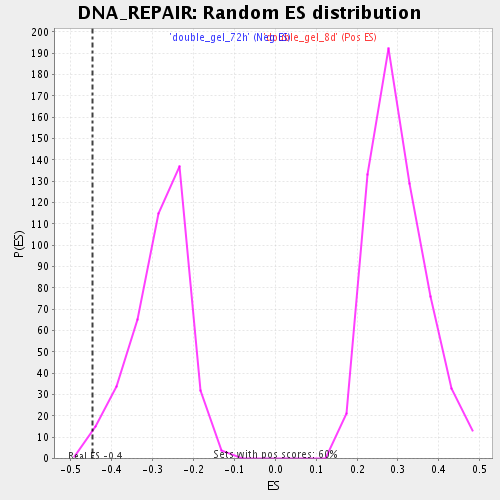
| Enrichment Score (ES) | -0.44694984 |
| Normalized Enrichment Score (NES) | -1.5824816 |
| Nominal p-value | 0.004962779 |
| FDR q-value | 0.21032149 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 326 | 0.238 | 0.0325 | No |
| 2 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 586 | 0.167 | 0.0530 | No |
| 3 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 1582 | 0.090 | -0.0004 | No |
| 4 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 1606 | 0.089 | 0.0192 | No |
| 5 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 2135 | 0.069 | -0.0040 | No |
| 6 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 2991 | 0.049 | -0.0567 | No |
| 7 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 3170 | 0.045 | -0.0593 | No |
| 8 | CIB1 | CIB1 Entrez, Source | calcium and integrin binding 1 (calmyrin) | 3206 | 0.045 | -0.0512 | No |
| 9 | SUMO1 | SUMO1 Entrez, Source | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 3347 | 0.042 | -0.0518 | No |
| 10 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 3405 | 0.040 | -0.0464 | No |
| 11 | RAD23A | RAD23A Entrez, Source | RAD23 homolog A (S. cerevisiae) | 3908 | 0.032 | -0.0766 | No |
| 12 | CSNK1D | CSNK1D Entrez, Source | casein kinase 1, delta | 4393 | 0.024 | -0.1073 | No |
| 13 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 4556 | 0.022 | -0.1143 | No |
| 14 | HTATIP | HTATIP Entrez, Source | HIV-1 Tat interacting protein, 60kDa | 4633 | 0.021 | -0.1151 | No |
| 15 | FANCC | FANCC Entrez, Source | Fanconi anemia, complementation group C | 4880 | 0.017 | -0.1295 | No |
| 16 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 5284 | 0.011 | -0.1571 | No |
| 17 | NBN | NBN Entrez, Source | nibrin | 5769 | 0.005 | -0.1924 | No |
| 18 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 5788 | 0.005 | -0.1927 | No |
| 19 | CSNK1E | CSNK1E Entrez, Source | casein kinase 1, epsilon | 5823 | 0.004 | -0.1942 | No |
| 20 | TNP1 | TNP1 Entrez, Source | transition protein 1 (during histone to protamine replacement) | 6455 | -0.004 | -0.2408 | No |
| 21 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 6464 | -0.004 | -0.2404 | No |
| 22 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 6859 | -0.009 | -0.2680 | No |
| 23 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 6922 | -0.010 | -0.2703 | No |
| 24 | OGG1 | OGG1 Entrez, Source | 8-oxoguanine DNA glycosylase | 7030 | -0.011 | -0.2757 | No |
| 25 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 7272 | -0.014 | -0.2905 | No |
| 26 | VCP | VCP Entrez, Source | valosin-containing protein | 7423 | -0.016 | -0.2981 | No |
| 27 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 7961 | -0.023 | -0.3330 | No |
| 28 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7962 | -0.023 | -0.3275 | No |
| 29 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 8325 | -0.028 | -0.3480 | No |
| 30 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 8535 | -0.031 | -0.3562 | No |
| 31 | RECQL | RECQL Entrez, Source | RecQ protein-like (DNA helicase Q1-like) | 8536 | -0.031 | -0.3487 | No |
| 32 | UBE2A | UBE2A Entrez, Source | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | 8758 | -0.034 | -0.3571 | No |
| 33 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 8883 | -0.036 | -0.3578 | No |
| 34 | DDB1 | DDB1 Entrez, Source | damage-specific DNA binding protein 1, 127kDa | 9492 | -0.045 | -0.3928 | No |
| 35 | IGHMBP2 | IGHMBP2 Entrez, Source | immunoglobulin mu binding protein 2 | 10212 | -0.057 | -0.4333 | Yes |
| 36 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 10227 | -0.057 | -0.4206 | Yes |
| 37 | MUTYH | MUTYH Entrez, Source | mutY homolog (E. coli) | 10230 | -0.057 | -0.4071 | Yes |
| 38 | UBE2V2 | UBE2V2 Entrez, Source | ubiquitin-conjugating enzyme E2 variant 2 | 10326 | -0.059 | -0.4001 | Yes |
| 39 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 10456 | -0.061 | -0.3951 | Yes |
| 40 | ATXN3 | ATXN3 Entrez, Source | ataxin 3 | 10628 | -0.065 | -0.3924 | Yes |
| 41 | MPG | MPG Entrez, Source | N-methylpurine-DNA glycosylase | 10650 | -0.065 | -0.3784 | Yes |
| 42 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 10927 | -0.072 | -0.3820 | Yes |
| 43 | WRNIP1 | WRNIP1 Entrez, Source | Werner helicase interacting protein 1 | 11093 | -0.076 | -0.3763 | Yes |
| 44 | PNKP | PNKP Entrez, Source | polynucleotide kinase 3'-phosphatase | 11117 | -0.076 | -0.3597 | Yes |
| 45 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 11274 | -0.081 | -0.3522 | Yes |
| 46 | XAB2 | XAB2 Entrez, Source | XPA binding protein 2 | 11397 | -0.084 | -0.3412 | Yes |
| 47 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 11496 | -0.086 | -0.3280 | Yes |
| 48 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 11628 | -0.090 | -0.3163 | Yes |
| 49 | GTF2H4 | GTF2H4 Entrez, Source | general transcription factor IIH, polypeptide 4, 52kDa | 12316 | -0.117 | -0.3400 | Yes |
| 50 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 12418 | -0.122 | -0.3183 | Yes |
| 51 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 12718 | -0.143 | -0.3065 | Yes |
| 52 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 12731 | -0.144 | -0.2729 | Yes |
| 53 | MSH3 | MSH3 Entrez, Source | mutS homolog 3 (E. coli) | 12763 | -0.148 | -0.2397 | Yes |
| 54 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 12780 | -0.150 | -0.2050 | Yes |
| 55 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 12879 | -0.162 | -0.1735 | Yes |
| 56 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 13022 | -0.190 | -0.1387 | Yes |
| 57 | APTX | APTX Entrez, Source | aprataxin | 13219 | -0.270 | -0.0886 | Yes |
| 58 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13302 | -0.408 | 0.0030 | Yes |