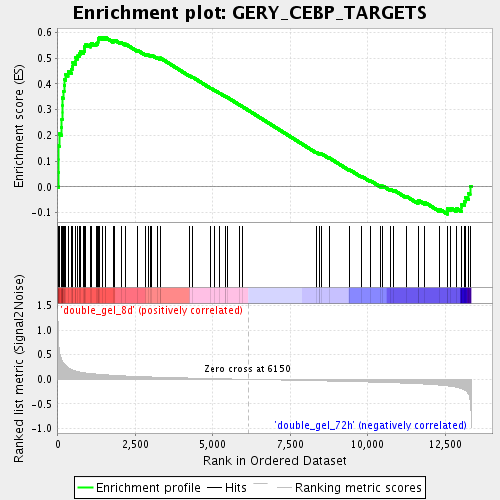
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | GERY_CEBP_TARGETS |
| Enrichment Score (ES) | 0.5811468 |
| Normalized Enrichment Score (NES) | 2.0432198 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0035258385 |
| FWER p-Value | 0.101 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DBP | DBP Entrez, Source | D site of albumin promoter (albumin D-box) binding protein | 16 | 0.727 | 0.0552 | Yes |
| 2 | LPL | LPL Entrez, Source | lipoprotein lipase | 24 | 0.680 | 0.1074 | Yes |
| 3 | PROCR | PROCR Entrez, Source | protein C receptor, endothelial (EPCR) | 26 | 0.669 | 0.1592 | Yes |
| 4 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 42 | 0.616 | 0.2059 | Yes |
| 5 | TNFAIP6 | TNFAIP6 Entrez, Source | tumor necrosis factor, alpha-induced protein 6 | 117 | 0.402 | 0.2315 | Yes |
| 6 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 126 | 0.391 | 0.2612 | Yes |
| 7 | CRYAB | CRYAB Entrez, Source | crystallin, alpha B | 141 | 0.371 | 0.2890 | Yes |
| 8 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 142 | 0.371 | 0.3177 | Yes |
| 9 | ANGPTL4 | ANGPTL4 Entrez, Source | angiopoietin-like 4 | 147 | 0.365 | 0.3457 | Yes |
| 10 | PRNP | PRNP Entrez, Source | prion protein (p27-30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 171 | 0.348 | 0.3709 | Yes |
| 11 | PLAT | PLAT Entrez, Source | plasminogen activator, tissue | 200 | 0.321 | 0.3937 | Yes |
| 12 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 218 | 0.309 | 0.4164 | Yes |
| 13 | PEA15 | PEA15 Entrez, Source | phosphoprotein enriched in astrocytes 15 | 252 | 0.281 | 0.4357 | Yes |
| 14 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 326 | 0.238 | 0.4487 | Yes |
| 15 | DNAJB9 | DNAJB9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 9 | 437 | 0.199 | 0.4558 | Yes |
| 16 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 459 | 0.193 | 0.4692 | Yes |
| 17 | IGF2 | IGF2 Entrez, Source | insulin-like growth factor 2 (somatomedin A) | 480 | 0.186 | 0.4821 | Yes |
| 18 | PER2 | PER2 Entrez, Source | period homolog 2 (Drosophila) | 566 | 0.171 | 0.4889 | Yes |
| 19 | BAIAP2 | BAIAP2 Entrez, Source | BAI1-associated protein 2 | 572 | 0.169 | 0.5016 | Yes |
| 20 | ZFAND2A | ZFAND2A Entrez, Source | zinc finger, AN1-type domain 2A | 644 | 0.158 | 0.5085 | Yes |
| 21 | NR1D1 | NR1D1 Entrez, Source | nuclear receptor subfamily 1, group D, member 1 | 688 | 0.151 | 0.5170 | Yes |
| 22 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 717 | 0.149 | 0.5265 | Yes |
| 23 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 841 | 0.135 | 0.5277 | Yes |
| 24 | APBB1IP | APBB1IP Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | 859 | 0.133 | 0.5367 | Yes |
| 25 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 866 | 0.132 | 0.5465 | Yes |
| 26 | MAFK | MAFK Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) | 900 | 0.129 | 0.5541 | Yes |
| 27 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1045 | 0.118 | 0.5523 | Yes |
| 28 | KLF4 | KLF4 Entrez, Source | Kruppel-like factor 4 (gut) | 1086 | 0.116 | 0.5583 | Yes |
| 29 | RASL11B | RASL11B Entrez, Source | RAS-like, family 11, member B | 1232 | 0.108 | 0.5557 | Yes |
| 30 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1289 | 0.105 | 0.5596 | Yes |
| 31 | IER2 | IER2 Entrez, Source | immediate early response 2 | 1295 | 0.104 | 0.5674 | Yes |
| 32 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 1297 | 0.104 | 0.5754 | Yes |
| 33 | PLAUR | PLAUR Entrez, Source | plasminogen activator, urokinase receptor | 1327 | 0.103 | 0.5811 | Yes |
| 34 | ACTA2 | ACTA2 Entrez, Source | actin, alpha 2, smooth muscle, aorta | 1439 | 0.096 | 0.5802 | No |
| 35 | CXCL1 | CXCL1 Entrez, Source | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 1541 | 0.092 | 0.5798 | No |
| 36 | S100A3 | S100A3 Entrez, Source | S100 calcium binding protein A3 | 1782 | 0.081 | 0.5679 | No |
| 37 | CXCL5 | CXCL5 Entrez, Source | chemokine (C-X-C motif) ligand 5 | 1832 | 0.079 | 0.5704 | No |
| 38 | MYD116 | 2037 | 0.072 | 0.5606 | No | ||
| 39 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 2181 | 0.068 | 0.5550 | No |
| 40 | CRABP2 | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 2566 | 0.058 | 0.5306 | No |
| 41 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 2829 | 0.052 | 0.5149 | No |
| 42 | MKNK2 | MKNK2 Entrez, Source | MAP kinase interacting serine/threonine kinase 2 | 2914 | 0.051 | 0.5124 | No |
| 43 | EREG | EREG Entrez, Source | epiregulin | 2999 | 0.049 | 0.5099 | No |
| 44 | DCN | DCN Entrez, Source | decorin | 3029 | 0.048 | 0.5114 | No |
| 45 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 3216 | 0.044 | 0.5008 | No |
| 46 | CLU | CLU Entrez, Source | clusterin | 3299 | 0.043 | 0.4979 | No |
| 47 | NRP1 | NRP1 Entrez, Source | neuropilin 1 | 3304 | 0.042 | 0.5009 | No |
| 48 | S100A8 | S100A8 Entrez, Source | S100 calcium binding protein A8 | 4260 | 0.026 | 0.4309 | No |
| 49 | FOXG1 | 4358 | 0.024 | 0.4255 | No | ||
| 50 | LCN2 | LCN2 Entrez, Source | lipocalin 2 (oncogene 24p3) | 4934 | 0.016 | 0.3834 | No |
| 51 | CD47 | CD47 Entrez, Source | CD47 molecule | 5066 | 0.014 | 0.3746 | No |
| 52 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 5229 | 0.012 | 0.3633 | No |
| 53 | SERPINI1 | SERPINI1 Entrez, Source | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 5393 | 0.010 | 0.3518 | No |
| 54 | HP | HP Entrez, Source | haptoglobin | 5419 | 0.009 | 0.3506 | No |
| 55 | BAG3 | BAG3 Entrez, Source | BCL2-associated athanogene 3 | 5477 | 0.009 | 0.3470 | No |
| 56 | FGF7 | FGF7 Entrez, Source | fibroblast growth factor 7 (keratinocyte growth factor) | 5875 | 0.004 | 0.3173 | No |
| 57 | ORM1 | ORM1 Entrez, Source | orosomucoid 1 | 5969 | 0.002 | 0.3105 | No |
| 58 | SERPINB2 | SERPINB2 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 8340 | -0.028 | 0.1340 | No |
| 59 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 8441 | -0.030 | 0.1288 | No |
| 60 | NR2F6 | NR2F6 Entrez, Source | nuclear receptor subfamily 2, group F, member 6 | 8452 | -0.030 | 0.1303 | No |
| 61 | PPARG | PPARG Entrez, Source | peroxisome proliferative activated receptor, gamma | 8513 | -0.031 | 0.1282 | No |
| 62 | C3 | C3 Entrez, Source | complement component 3 | 8749 | -0.034 | 0.1131 | No |
| 63 | RRAD | RRAD Entrez, Source | Ras-related associated with diabetes | 9408 | -0.044 | 0.0669 | No |
| 64 | FHL2 | FHL2 Entrez, Source | four and a half LIM domains 2 | 9790 | -0.050 | 0.0420 | No |
| 65 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 10096 | -0.055 | 0.0233 | No |
| 66 | CSPG2 | CSPG2 Entrez, Source | chondroitin sulfate proteoglycan 2 (versican) | 10417 | -0.061 | 0.0039 | No |
| 67 | HGF | HGF Entrez, Source | hepatocyte growth factor (hepapoietin A; scatter factor) | 10486 | -0.062 | 0.0035 | No |
| 68 | RNH1 | RNH1 Entrez, Source | ribonuclease/angiogenin inhibitor 1 | 10745 | -0.067 | -0.0107 | No |
| 69 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 10842 | -0.069 | -0.0126 | No |
| 70 | CD1D1 | 11237 | -0.079 | -0.0361 | No | ||
| 71 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 11624 | -0.090 | -0.0583 | No |
| 72 | TRIB3 | TRIB3 Entrez, Source | tribbles homolog 3 (Drosophila) | 11637 | -0.090 | -0.0522 | No |
| 73 | HMGCR | HMGCR Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 11844 | -0.097 | -0.0602 | No |
| 74 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 12319 | -0.117 | -0.0869 | No |
| 75 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 12570 | -0.133 | -0.0954 | No |
| 76 | CD68 | CD68 Entrez, Source | CD68 molecule | 12573 | -0.133 | -0.0853 | No |
| 77 | MVD | MVD Entrez, Source | mevalonate (diphospho) decarboxylase | 12678 | -0.140 | -0.0823 | No |
| 78 | LSS | LSS Entrez, Source | lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) | 12854 | -0.159 | -0.0832 | No |
| 79 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 13029 | -0.193 | -0.0813 | No |
| 80 | SH3GL3 | SH3GL3 Entrez, Source | SH3-domain GRB2-like 3 | 13033 | -0.194 | -0.0666 | No |
| 81 | XDH | XDH Entrez, Source | xanthine dehydrogenase | 13126 | -0.217 | -0.0566 | No |
| 82 | DDIT3 | DDIT3 Entrez, Source | DNA-damage-inducible transcript 3 | 13156 | -0.227 | -0.0412 | No |
| 83 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 13254 | -0.302 | -0.0251 | No |
| 84 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13302 | -0.408 | 0.0030 | No |

