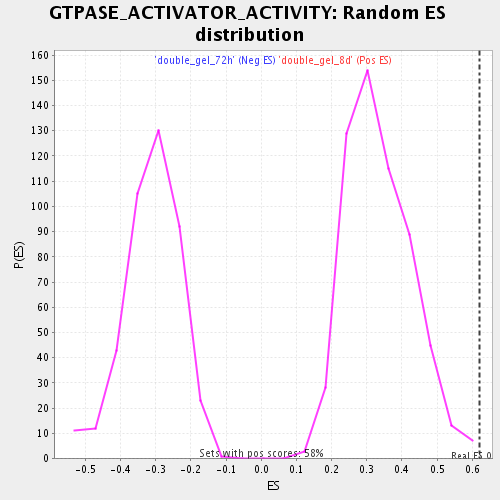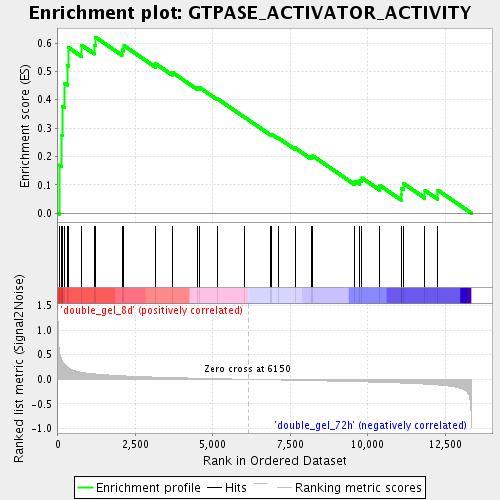
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | GTPASE_ACTIVATOR_ACTIVITY |
| Enrichment Score (ES) | 0.6202941 |
| Normalized Enrichment Score (NES) | 1.8506734 |
| Nominal p-value | 0.0017152659 |
| FDR q-value | 0.023180347 |
| FWER p-Value | 0.854 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 42 | 0.616 | 0.1699 | Yes |
| 2 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 126 | 0.391 | 0.2737 | Yes |
| 3 | RGS1 | RGS1 Entrez, Source | regulator of G-protein signalling 1 | 140 | 0.372 | 0.3774 | Yes |
| 4 | RGS4 | RGS4 Entrez, Source | regulator of G-protein signalling 4 | 219 | 0.308 | 0.4582 | Yes |
| 5 | RGS5 | RGS5 Entrez, Source | regulator of G-protein signalling 5 | 306 | 0.247 | 0.5211 | Yes |
| 6 | CDC42EP2 | CDC42EP2 Entrez, Source | CDC42 effector protein (Rho GTPase binding) 2 | 343 | 0.233 | 0.5838 | Yes |
| 7 | RGS3 | RGS3 Entrez, Source | regulator of G-protein signalling 3 | 762 | 0.144 | 0.5927 | Yes |
| 8 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 1183 | 0.110 | 0.5922 | Yes |
| 9 | RASA2 | RASA2 Entrez, Source | RAS p21 protein activator 2 | 1216 | 0.109 | 0.6203 | Yes |
| 10 | ERRFI1 | ERRFI1 Entrez, Source | ERBB receptor feedback inhibitor 1 | 2071 | 0.071 | 0.5760 | No |
| 11 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 2131 | 0.069 | 0.5910 | No |
| 12 | RAP1GAP | RAP1GAP Entrez, Source | RAP1 GTPase activating protein | 3159 | 0.045 | 0.5266 | No |
| 13 | RALBP1 | RALBP1 Entrez, Source | ralA binding protein 1 | 3689 | 0.035 | 0.4968 | No |
| 14 | CENTA1 | CENTA1 Entrez, Source | centaurin, alpha 1 | 4506 | 0.022 | 0.4417 | No |
| 15 | RGS12 | RGS12 Entrez, Source | regulator of G-protein signalling 12 | 4561 | 0.022 | 0.4438 | No |
| 16 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 5138 | 0.013 | 0.4042 | No |
| 17 | RGS9 | RGS9 Entrez, Source | regulator of G-protein signalling 9 | 6017 | 0.002 | 0.3388 | No |
| 18 | RGS14 | RGS14 Entrez, Source | regulator of G-protein signalling 14 | 6865 | -0.009 | 0.2777 | No |
| 19 | ARHGAP27 | ARHGAP27 Entrez, Source | Rho GTPase activating protein 27 | 6907 | -0.010 | 0.2773 | No |
| 20 | ARHGAP4 | ARHGAP4 Entrez, Source | Rho GTPase activating protein 4 | 7114 | -0.012 | 0.2652 | No |
| 21 | RGS6 | RGS6 Entrez, Source | regulator of G-protein signalling 6 | 7652 | -0.019 | 0.2302 | No |
| 22 | RANGAP1 | RANGAP1 Entrez, Source | Ran GTPase activating protein 1 | 8169 | -0.026 | 0.1988 | No |
| 23 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 8226 | -0.027 | 0.2021 | No |
| 24 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 9586 | -0.047 | 0.1131 | No |
| 25 | MYO9B | MYO9B Entrez, Source | myosin IXB | 9740 | -0.049 | 0.1154 | No |
| 26 | TSC2 | TSC2 Entrez, Source | tuberous sclerosis 2 | 9813 | -0.050 | 0.1242 | No |
| 27 | ARHGAP5 | ARHGAP5 Entrez, Source | Rho GTPase activating protein 5 | 10392 | -0.060 | 0.0976 | No |
| 28 | CHN2 | CHN2 Entrez, Source | chimerin (chimaerin) 2 | 11079 | -0.075 | 0.0672 | No |
| 29 | RASA3 | RASA3 Entrez, Source | RAS p21 protein activator 3 | 11084 | -0.075 | 0.0881 | No |
| 30 | RAB3GAP2 | RAB3GAP2 Entrez, Source | RAB3 GTPase activating protein subunit 2 (non-catalytic) | 11166 | -0.077 | 0.1037 | No |
| 31 | CENTD2 | CENTD2 Entrez, Source | centaurin, delta 2 | 11845 | -0.097 | 0.0802 | No |
| 32 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 12263 | -0.115 | 0.0811 | No |