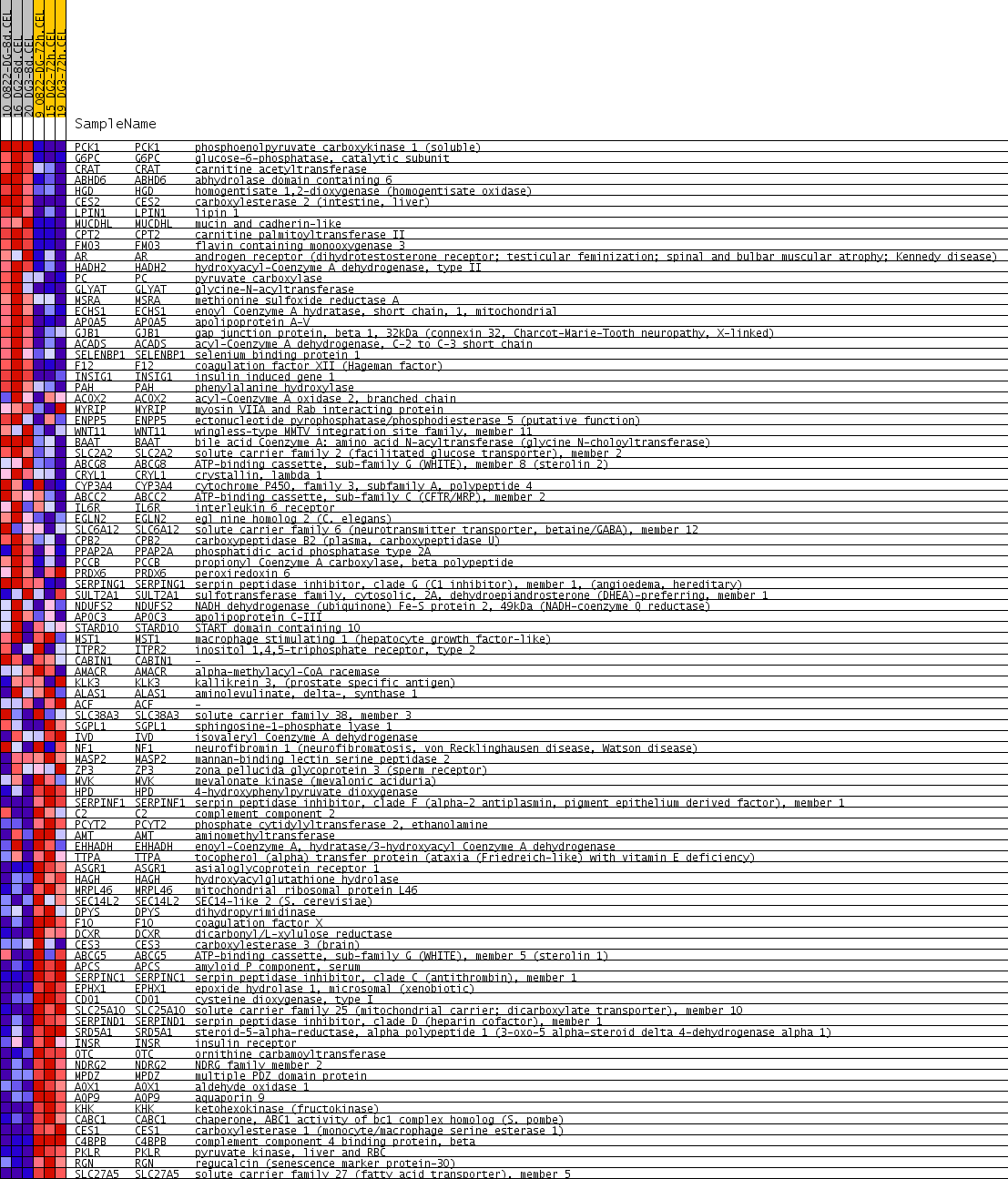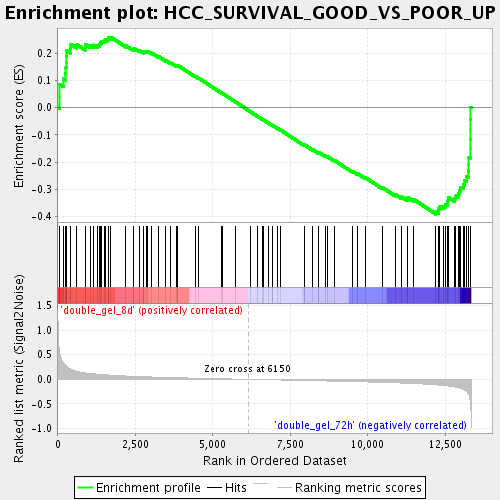
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HCC_SURVIVAL_GOOD_VS_POOR_UP |
| Enrichment Score (ES) | -0.39097875 |
| Normalized Enrichment Score (NES) | -1.4946576 |
| Nominal p-value | 0.0077120825 |
| FDR q-value | 0.32977936 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 59 | 0.557 | 0.0409 | No |
| 2 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 60 | 0.555 | 0.0861 | No |
| 3 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 170 | 0.348 | 0.1062 | No |
| 4 | ABHD6 | ABHD6 Entrez, Source | abhydrolase domain containing 6 | 235 | 0.296 | 0.1255 | No |
| 5 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 258 | 0.276 | 0.1463 | No |
| 6 | CES2 | CES2 Entrez, Source | carboxylesterase 2 (intestine, liver) | 265 | 0.270 | 0.1679 | No |
| 7 | LPIN1 | LPIN1 Entrez, Source | lipin 1 | 269 | 0.266 | 0.1893 | No |
| 8 | MUCDHL | MUCDHL Entrez, Source | mucin and cadherin-like | 285 | 0.256 | 0.2091 | No |
| 9 | CPT2 | CPT2 Entrez, Source | carnitine palmitoyltransferase II | 399 | 0.209 | 0.2176 | No |
| 10 | FMO3 | FMO3 Entrez, Source | flavin containing monooxygenase 3 | 416 | 0.205 | 0.2331 | No |
| 11 | AR | AR Entrez, Source | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; Kennedy disease) | 602 | 0.164 | 0.2325 | No |
| 12 | HADH2 | HADH2 Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase, type II | 876 | 0.131 | 0.2226 | No |
| 13 | PC | PC Entrez, Source | pyruvate carboxylase | 895 | 0.130 | 0.2318 | No |
| 14 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 1056 | 0.117 | 0.2293 | No |
| 15 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1160 | 0.111 | 0.2306 | No |
| 16 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1289 | 0.105 | 0.2294 | No |
| 17 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 1344 | 0.102 | 0.2336 | No |
| 18 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 1380 | 0.100 | 0.2391 | No |
| 19 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1417 | 0.097 | 0.2443 | No |
| 20 | SELENBP1 | SELENBP1 Entrez, Source | selenium binding protein 1 | 1492 | 0.094 | 0.2464 | No |
| 21 | F12 | F12 Entrez, Source | coagulation factor XII (Hageman factor) | 1525 | 0.093 | 0.2515 | No |
| 22 | INSIG1 | INSIG1 Entrez, Source | insulin induced gene 1 | 1616 | 0.089 | 0.2520 | No |
| 23 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 1638 | 0.087 | 0.2575 | No |
| 24 | ACOX2 | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 1711 | 0.084 | 0.2589 | No |
| 25 | MYRIP | MYRIP Entrez, Source | myosin VIIA and Rab interacting protein | 2192 | 0.067 | 0.2282 | No |
| 26 | ENPP5 | ENPP5 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative function) | 2442 | 0.061 | 0.2143 | No |
| 27 | WNT11 | WNT11 Entrez, Source | wingless-type MMTV integration site family, member 11 | 2454 | 0.061 | 0.2185 | No |
| 28 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 2620 | 0.057 | 0.2107 | No |
| 29 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 2754 | 0.054 | 0.2050 | No |
| 30 | ABCG8 | ABCG8 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 8 (sterolin 2) | 2769 | 0.053 | 0.2083 | No |
| 31 | CRYL1 | CRYL1 Entrez, Source | crystallin, lambda 1 | 2849 | 0.052 | 0.2065 | No |
| 32 | CYP3A4 | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 2896 | 0.051 | 0.2072 | No |
| 33 | ABCC2 | ABCC2 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 3028 | 0.048 | 0.2012 | No |
| 34 | IL6R | IL6R Entrez, Source | interleukin 6 receptor | 3241 | 0.044 | 0.1888 | No |
| 35 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 3481 | 0.039 | 0.1739 | No |
| 36 | SLC6A12 | SLC6A12 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 | 3625 | 0.036 | 0.1661 | No |
| 37 | CPB2 | CPB2 Entrez, Source | carboxypeptidase B2 (plasma, carboxypeptidase U) | 3831 | 0.033 | 0.1533 | No |
| 38 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 3851 | 0.033 | 0.1545 | No |
| 39 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 3873 | 0.032 | 0.1555 | No |
| 40 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 4430 | 0.023 | 0.1155 | No |
| 41 | SERPING1 | SERPING1 Entrez, Source | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) | 4547 | 0.022 | 0.1085 | No |
| 42 | SULT2A1 | SULT2A1 Entrez, Source | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 | 5283 | 0.011 | 0.0539 | No |
| 43 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 5319 | 0.011 | 0.0522 | No |
| 44 | APOC3 | APOC3 Entrez, Source | apolipoprotein C-III | 5734 | 0.005 | 0.0214 | No |
| 45 | STARD10 | STARD10 Entrez, Source | START domain containing 10 | 6223 | -0.001 | -0.0154 | No |
| 46 | MST1 | MST1 Entrez, Source | macrophage stimulating 1 (hepatocyte growth factor-like) | 6456 | -0.004 | -0.0326 | No |
| 47 | ITPR2 | ITPR2 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 2 | 6591 | -0.006 | -0.0422 | No |
| 48 | CABIN1 | CABIN1 Entrez, Source | - | 6635 | -0.006 | -0.0450 | No |
| 49 | AMACR | AMACR Entrez, Source | alpha-methylacyl-CoA racemase | 6811 | -0.008 | -0.0575 | No |
| 50 | KLK3 | KLK3 Entrez, Source | kallikrein 3, (prostate specific antigen) | 6910 | -0.010 | -0.0641 | No |
| 51 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 6928 | -0.010 | -0.0646 | No |
| 52 | ACF | ACF Entrez, Source | - | 7081 | -0.012 | -0.0751 | No |
| 53 | SLC38A3 | SLC38A3 Entrez, Source | solute carrier family 38, member 3 | 7186 | -0.013 | -0.0819 | No |
| 54 | SGPL1 | SGPL1 Entrez, Source | sphingosine-1-phosphate lyase 1 | 7952 | -0.023 | -0.1378 | No |
| 55 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 7956 | -0.023 | -0.1362 | No |
| 56 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 8226 | -0.027 | -0.1543 | No |
| 57 | MASP2 | MASP2 Entrez, Source | mannan-binding lectin serine peptidase 2 | 8407 | -0.029 | -0.1655 | No |
| 58 | ZP3 | ZP3 Entrez, Source | zona pellucida glycoprotein 3 (sperm receptor) | 8416 | -0.030 | -0.1637 | No |
| 59 | MVK | MVK Entrez, Source | mevalonate kinase (mevalonic aciduria) | 8632 | -0.033 | -0.1773 | No |
| 60 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 8712 | -0.034 | -0.1805 | No |
| 61 | SERPINF1 | SERPINF1 Entrez, Source | serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 | 8926 | -0.036 | -0.1936 | No |
| 62 | C2 | C2 Entrez, Source | complement component 2 | 9498 | -0.045 | -0.2330 | No |
| 63 | PCYT2 | PCYT2 Entrez, Source | phosphate cytidylyltransferase 2, ethanolamine | 9674 | -0.048 | -0.2423 | No |
| 64 | AMT | AMT Entrez, Source | aminomethyltransferase | 9912 | -0.052 | -0.2559 | No |
| 65 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 10463 | -0.061 | -0.2925 | No |
| 66 | TTPA | TTPA Entrez, Source | tocopherol (alpha) transfer protein (ataxia (Friedreich-like) with vitamin E deficiency) | 10904 | -0.071 | -0.3199 | No |
| 67 | ASGR1 | ASGR1 Entrez, Source | asialoglycoprotein receptor 1 | 11077 | -0.075 | -0.3268 | No |
| 68 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 11283 | -0.081 | -0.3357 | No |
| 69 | MRPL46 | MRPL46 Entrez, Source | mitochondrial ribosomal protein L46 | 11292 | -0.081 | -0.3297 | No |
| 70 | SEC14L2 | SEC14L2 Entrez, Source | SEC14-like 2 (S. cerevisiae) | 11482 | -0.086 | -0.3369 | No |
| 71 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 12199 | -0.111 | -0.3819 | Yes |
| 72 | F10 | F10 Entrez, Source | coagulation factor X | 12278 | -0.115 | -0.3784 | Yes |
| 73 | DCXR | DCXR Entrez, Source | dicarbonyl/L-xylulose reductase | 12287 | -0.116 | -0.3696 | Yes |
| 74 | CES3 | CES3 Entrez, Source | carboxylesterase 3 (brain) | 12329 | -0.117 | -0.3632 | Yes |
| 75 | ABCG5 | ABCG5 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 5 (sterolin 1) | 12442 | -0.123 | -0.3616 | Yes |
| 76 | APCS | APCS Entrez, Source | amyloid P component, serum | 12502 | -0.128 | -0.3556 | Yes |
| 77 | SERPINC1 | SERPINC1 Entrez, Source | serpin peptidase inhibitor, clade C (antithrombin), member 1 | 12565 | -0.133 | -0.3495 | Yes |
| 78 | EPHX1 | EPHX1 Entrez, Source | epoxide hydrolase 1, microsomal (xenobiotic) | 12587 | -0.134 | -0.3402 | Yes |
| 79 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 12594 | -0.134 | -0.3297 | Yes |
| 80 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 12799 | -0.152 | -0.3327 | Yes |
| 81 | SERPIND1 | SERPIND1 Entrez, Source | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | 12846 | -0.158 | -0.3233 | Yes |
| 82 | SRD5A1 | SRD5A1 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 12915 | -0.167 | -0.3148 | Yes |
| 83 | INSR | INSR Entrez, Source | insulin receptor | 12948 | -0.173 | -0.3032 | Yes |
| 84 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 12994 | -0.182 | -0.2917 | Yes |
| 85 | NDRG2 | NDRG2 Entrez, Source | NDRG family member 2 | 13082 | -0.204 | -0.2816 | Yes |
| 86 | MPDZ | MPDZ Entrez, Source | multiple PDZ domain protein | 13135 | -0.221 | -0.2676 | Yes |
| 87 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 13183 | -0.242 | -0.2514 | Yes |
| 88 | AQP9 | AQP9 Entrez, Source | aquaporin 9 | 13248 | -0.296 | -0.2321 | Yes |
| 89 | KHK | KHK Entrez, Source | ketohexokinase (fructokinase) | 13261 | -0.313 | -0.2075 | Yes |
| 90 | CABC1 | CABC1 Entrez, Source | chaperone, ABC1 activity of bc1 complex homolog (S. pombe) | 13264 | -0.315 | -0.1820 | Yes |
| 91 | CES1 | CES1 Entrez, Source | carboxylesterase 1 (monocyte/macrophage serine esterase 1) | 13305 | -0.420 | -0.1509 | Yes |
| 92 | C4BPB | C4BPB Entrez, Source | complement component 4 binding protein, beta | 13307 | -0.428 | -0.1161 | Yes |
| 93 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 13313 | -0.446 | -0.0801 | Yes |
| 94 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13314 | -0.464 | -0.0424 | Yes |
| 95 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 13326 | -0.545 | 0.0012 | Yes |