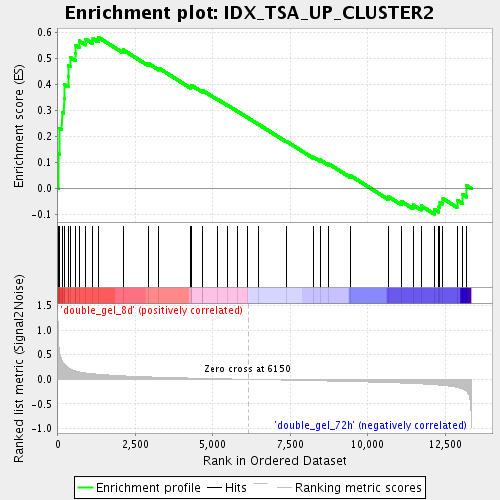
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | IDX_TSA_UP_CLUSTER2 |
| Enrichment Score (ES) | 0.5805955 |
| Normalized Enrichment Score (NES) | 1.7878678 |
| Nominal p-value | 0.0017793594 |
| FDR q-value | 0.03333546 |
| FWER p-Value | 0.979 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL1RL1 | IL1RL1 Entrez, Source | interleukin 1 receptor-like 1 | 13 | 0.781 | 0.1343 | Yes |
| 2 | COL4A1 | COL4A1 Entrez, Source | collagen, type IV, alpha 1 | 50 | 0.578 | 0.2318 | Yes |
| 3 | FST | FST Entrez, Source | follistatin | 132 | 0.388 | 0.2929 | Yes |
| 4 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 196 | 0.324 | 0.3443 | Yes |
| 5 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 201 | 0.321 | 0.3996 | Yes |
| 6 | RAMP3 | RAMP3 Entrez, Source | receptor (calcitonin) activity modifying protein 3 | 329 | 0.237 | 0.4311 | Yes |
| 7 | FKBP5 | FKBP5 Entrez, Source | FK506 binding protein 5 | 335 | 0.235 | 0.4714 | Yes |
| 8 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 396 | 0.210 | 0.5033 | Yes |
| 9 | PER2 | PER2 Entrez, Source | period homolog 2 (Drosophila) | 566 | 0.171 | 0.5201 | Yes |
| 10 | TGFBR3 | TGFBR3 Entrez, Source | transforming growth factor, beta receptor III (betaglycan, 300kDa) | 577 | 0.168 | 0.5485 | Yes |
| 11 | SAR1B | SAR1B Entrez, Source | SAR1 gene homolog B (S. cerevisiae) | 686 | 0.152 | 0.5667 | Yes |
| 12 | LIMK1 | LIMK1 Entrez, Source | LIM domain kinase 1 | 899 | 0.129 | 0.5732 | Yes |
| 13 | SNAI1 | SNAI1 Entrez, Source | snail homolog 1 (Drosophila) | 1130 | 0.113 | 0.5755 | Yes |
| 14 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 1303 | 0.104 | 0.5806 | Yes |
| 15 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 2106 | 0.069 | 0.5323 | No |
| 16 | MKNK2 | MKNK2 Entrez, Source | MAP kinase interacting serine/threonine kinase 2 | 2914 | 0.051 | 0.4804 | No |
| 17 | TIMP1 | TIMP1 Entrez, Source | TIMP metallopeptidase inhibitor 1 | 3256 | 0.044 | 0.4623 | No |
| 18 | TPM4 | TPM4 Entrez, Source | tropomyosin 4 | 4281 | 0.026 | 0.3898 | No |
| 19 | MCL1 | MCL1 Entrez, Source | myeloid cell leukemia sequence 1 (BCL2-related) | 4307 | 0.025 | 0.3923 | No |
| 20 | LXN | LXN Entrez, Source | latexin | 4313 | 0.025 | 0.3963 | No |
| 21 | IL1R1 | IL1R1 Entrez, Source | interleukin 1 receptor, type I | 4666 | 0.020 | 0.3733 | No |
| 22 | PTGES | PTGES Entrez, Source | prostaglandin E synthase | 4675 | 0.020 | 0.3761 | No |
| 23 | HSPD1 | HSPD1 Entrez, Source | heat shock 60kDa protein 1 (chaperonin) | 5153 | 0.013 | 0.3426 | No |
| 24 | NF2 | NF2 Entrez, Source | neurofibromin 2 (bilateral acoustic neuroma) | 5466 | 0.009 | 0.3206 | No |
| 25 | PRKCE | PRKCE Entrez, Source | protein kinase C, epsilon | 5796 | 0.005 | 0.2967 | No |
| 26 | HSPH1 | HSPH1 Entrez, Source | heat shock 105kDa/110kDa protein 1 | 6127 | 0.000 | 0.2719 | No |
| 27 | AVPI1 | AVPI1 Entrez, Source | arginine vasopressin-induced 1 | 6462 | -0.004 | 0.2475 | No |
| 28 | SERP1 | SERP1 Entrez, Source | - | 7376 | -0.015 | 0.1815 | No |
| 29 | RHOJ | RHOJ Entrez, Source | ras homolog gene family, member J | 8240 | -0.027 | 0.1213 | No |
| 30 | VDR | VDR Entrez, Source | vitamin D (1,25- dihydroxyvitamin D3) receptor | 8460 | -0.030 | 0.1101 | No |
| 31 | FTSJ3 | FTSJ3 Entrez, Source | FtsJ homolog 3 (E. coli) | 8738 | -0.034 | 0.0951 | No |
| 32 | ANK3 | ANK3 Entrez, Source | ankyrin 3, node of Ranvier (ankyrin G) | 9444 | -0.044 | 0.0498 | No |
| 33 | GRWD1 | GRWD1 Entrez, Source | glutamate-rich WD repeat containing 1 | 10659 | -0.065 | -0.0302 | No |
| 34 | RASA3 | RASA3 Entrez, Source | RAS p21 protein activator 3 | 11084 | -0.075 | -0.0490 | No |
| 35 | KCNN4 | KCNN4 Entrez, Source | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 11462 | -0.085 | -0.0626 | No |
| 36 | TUBB3 | TUBB3 Entrez, Source | tubulin, beta 3 | 11724 | -0.093 | -0.0661 | No |
| 37 | SRR | SRR Entrez, Source | serine racemase | 12146 | -0.109 | -0.0789 | No |
| 38 | SRXN1 | SRXN1 Entrez, Source | sulfiredoxin 1 homolog (S. cerevisiae) | 12296 | -0.116 | -0.0700 | No |
| 39 | GSTO1 | GSTO1 Entrez, Source | glutathione S-transferase omega 1 | 12328 | -0.117 | -0.0520 | No |
| 40 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 12425 | -0.123 | -0.0379 | No |
| 41 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 12882 | -0.162 | -0.0441 | No |
| 42 | CLIC4 | CLIC4 Entrez, Source | chloride intracellular channel 4 | 13070 | -0.202 | -0.0232 | No |
| 43 | SFPQ | SFPQ Entrez, Source | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | 13196 | -0.252 | 0.0110 | No |

