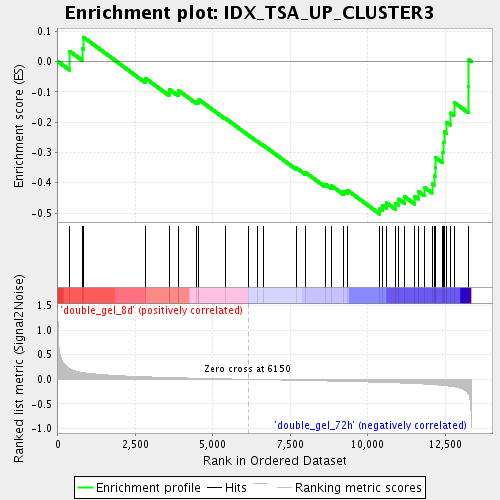
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
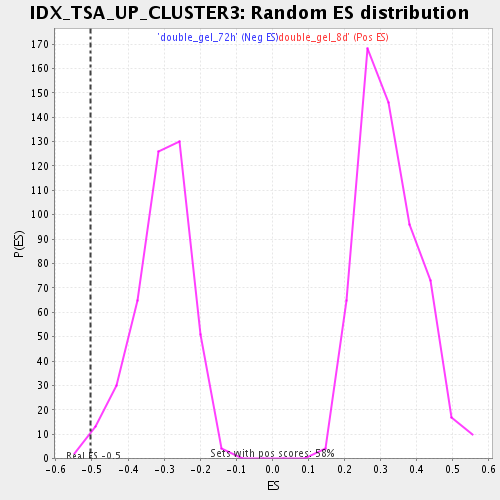
| GeneSet | IDX_TSA_UP_CLUSTER3 |
| Enrichment Score (ES) | -0.50333196 |
| Normalized Enrichment Score (NES) | -1.6463228 |
| Nominal p-value | 0.011876484 |
| FDR q-value | 0.19049086 |
| FWER p-Value | 1.0 |

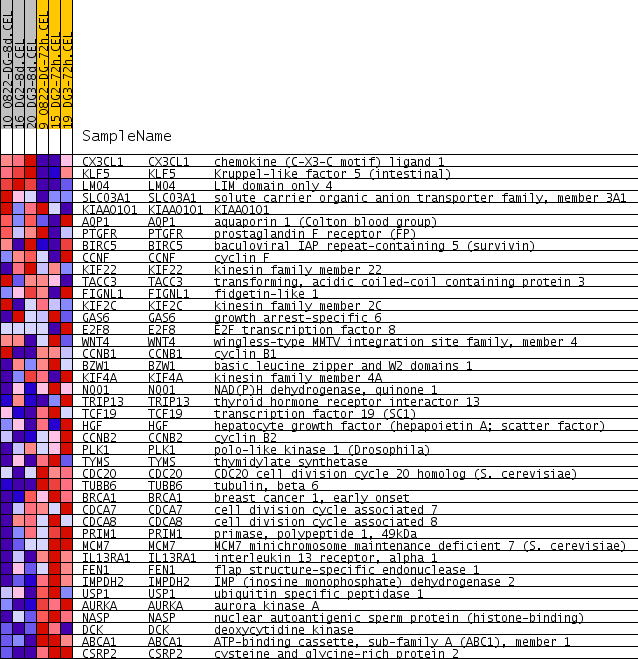
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CX3CL1 | CX3CL1 Entrez, Source | chemokine (C-X3-C motif) ligand 1 | 375 | 0.218 | 0.0342 | No |
| 2 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 780 | 0.141 | 0.0444 | No |
| 3 | LMO4 | LMO4 Entrez, Source | LIM domain only 4 | 827 | 0.136 | 0.0800 | No |
| 4 | SLCO3A1 | SLCO3A1 Entrez, Source | solute carrier organic anion transporter family, member 3A1 | 2830 | 0.052 | -0.0556 | No |
| 5 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 3593 | 0.037 | -0.1023 | No |
| 6 | AQP1 | AQP1 Entrez, Source | aquaporin 1 (Colton blood group) | 3608 | 0.037 | -0.0929 | No |
| 7 | PTGFR | PTGFR Entrez, Source | prostaglandin F receptor (FP) | 3885 | 0.032 | -0.1044 | No |
| 8 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 3898 | 0.032 | -0.0962 | No |
| 9 | CCNF | CCNF Entrez, Source | cyclin F | 4470 | 0.023 | -0.1326 | No |
| 10 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 4530 | 0.022 | -0.1308 | No |
| 11 | TACC3 | TACC3 Entrez, Source | transforming, acidic coiled-coil containing protein 3 | 4550 | 0.022 | -0.1259 | No |
| 12 | FIGNL1 | FIGNL1 Entrez, Source | fidgetin-like 1 | 5399 | 0.010 | -0.1869 | No |
| 13 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 6164 | -0.000 | -0.2443 | No |
| 14 | GAS6 | GAS6 Entrez, Source | growth arrest-specific 6 | 6432 | -0.004 | -0.2634 | No |
| 15 | E2F8 | E2F8 Entrez, Source | E2F transcription factor 8 | 6648 | -0.006 | -0.2777 | No |
| 16 | WNT4 | WNT4 Entrez, Source | wingless-type MMTV integration site family, member 4 | 7687 | -0.019 | -0.3502 | No |
| 17 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 8004 | -0.024 | -0.3672 | No |
| 18 | BZW1 | BZW1 Entrez, Source | basic leucine zipper and W2 domains 1 | 8626 | -0.033 | -0.4046 | No |
| 19 | KIF4A | KIF4A Entrez, Source | kinesin family member 4A | 8827 | -0.035 | -0.4095 | No |
| 20 | NQO1 | NQO1 Entrez, Source | NAD(P)H dehydrogenase, quinone 1 | 9220 | -0.040 | -0.4274 | No |
| 21 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 9335 | -0.042 | -0.4239 | No |
| 22 | TCF19 | TCF19 Entrez, Source | transcription factor 19 (SC1) | 10393 | -0.060 | -0.4861 | Yes |
| 23 | HGF | HGF Entrez, Source | hepatocyte growth factor (hepapoietin A; scatter factor) | 10486 | -0.062 | -0.4753 | Yes |
| 24 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 10615 | -0.065 | -0.4664 | Yes |
| 25 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 10899 | -0.071 | -0.4675 | Yes |
| 26 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 10993 | -0.073 | -0.4535 | Yes |
| 27 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11187 | -0.078 | -0.4457 | Yes |
| 28 | TUBB6 | TUBB6 Entrez, Source | tubulin, beta 6 | 11518 | -0.087 | -0.4456 | Yes |
| 29 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 11628 | -0.090 | -0.4281 | Yes |
| 30 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 11823 | -0.096 | -0.4150 | Yes |
| 31 | CDCA8 | CDCA8 Entrez, Source | cell division cycle associated 8 | 12074 | -0.106 | -0.4035 | Yes |
| 32 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12141 | -0.109 | -0.3773 | Yes |
| 33 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 12174 | -0.110 | -0.3480 | Yes |
| 34 | IL13RA1 | IL13RA1 Entrez, Source | interleukin 13 receptor, alpha 1 | 12191 | -0.111 | -0.3174 | Yes |
| 35 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 12418 | -0.122 | -0.2994 | Yes |
| 36 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 12451 | -0.124 | -0.2664 | Yes |
| 37 | USP1 | USP1 Entrez, Source | ubiquitin specific peptidase 1 | 12463 | -0.125 | -0.2314 | Yes |
| 38 | AURKA | AURKA Entrez, Source | aurora kinase A | 12546 | -0.131 | -0.1999 | Yes |
| 39 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 12677 | -0.140 | -0.1697 | Yes |
| 40 | DCK | DCK Entrez, Source | deoxycytidine kinase | 12784 | -0.151 | -0.1346 | Yes |
| 41 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 13254 | -0.302 | -0.0832 | Yes |
| 42 | CSRP2 | CSRP2 Entrez, Source | cysteine and glycine-rich protein 2 | 13262 | -0.313 | 0.0060 | Yes |