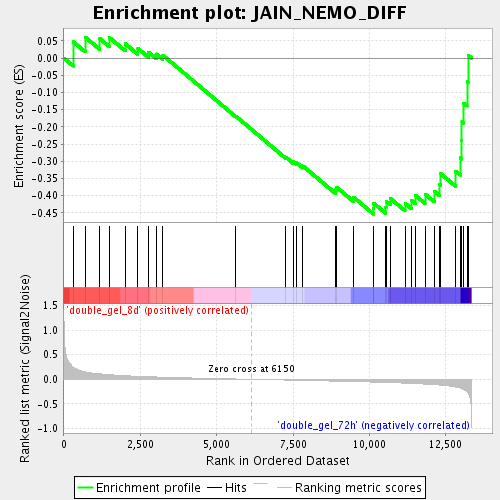
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | JAIN_NEMO_DIFF |
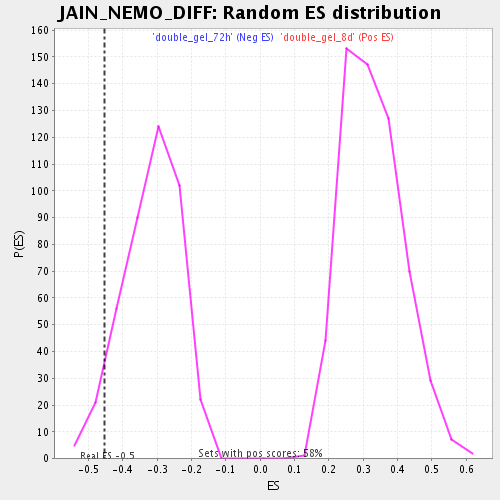
| Enrichment Score (ES) | -0.4537303 |
| Normalized Enrichment Score (NES) | -1.437345 |
| Nominal p-value | 0.054761905 |
| FDR q-value | 0.4269628 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DECR1 | DECR1 Entrez, Source | 2,4-dienoyl CoA reductase 1, mitochondrial | 304 | 0.247 | 0.0477 | No |
| 2 | FLT1 | FLT1 Entrez, Source | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 699 | 0.150 | 0.0609 | No |
| 3 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 1177 | 0.111 | 0.0565 | No |
| 4 | DCTN1 | DCTN1 Entrez, Source | dynactin 1 (p150, glued homolog, Drosophila) | 1493 | 0.094 | 0.0597 | No |
| 5 | PEX14 | PEX14 Entrez, Source | peroxisomal biogenesis factor 14 | 2007 | 0.073 | 0.0420 | No |
| 6 | PAWR | PAWR Entrez, Source | PRKC, apoptosis, WT1, regulator | 2421 | 0.061 | 0.0284 | No |
| 7 | ARSB | ARSB Entrez, Source | arylsulfatase B | 2773 | 0.053 | 0.0172 | No |
| 8 | SLC23A2 | SLC23A2 Entrez, Source | solute carrier family 23 (nucleobase transporters), member 2 | 3023 | 0.048 | 0.0122 | No |
| 9 | BCAT2 | BCAT2 Entrez, Source | branched chain aminotransferase 2, mitochondrial | 3239 | 0.044 | 0.0085 | No |
| 10 | HRSP12 | HRSP12 Entrez, Source | heat-responsive protein 12 | 5621 | 0.007 | -0.1685 | No |
| 11 | SYMPK | SYMPK Entrez, Source | symplekin | 7266 | -0.014 | -0.2881 | No |
| 12 | NAP1L1 | NAP1L1 Entrez, Source | nucleosome assembly protein 1-like 1 | 7512 | -0.017 | -0.3017 | No |
| 13 | PSMA2 | PSMA2 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 2 | 7610 | -0.018 | -0.3038 | No |
| 14 | HK2 | HK2 Entrez, Source | hexokinase 2 | 7823 | -0.021 | -0.3137 | No |
| 15 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 8883 | -0.036 | -0.3830 | No |
| 16 | SPHK2 | SPHK2 Entrez, Source | sphingosine kinase 2 | 8931 | -0.037 | -0.3761 | No |
| 17 | TPD52L2 | TPD52L2 Entrez, Source | tumor protein D52-like 2 | 9479 | -0.045 | -0.4045 | No |
| 18 | TAP2 | TAP2 Entrez, Source | transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | 10135 | -0.056 | -0.4379 | Yes |
| 19 | PTPN7 | PTPN7 Entrez, Source | protein tyrosine phosphatase, non-receptor type 7 | 10142 | -0.056 | -0.4224 | Yes |
| 20 | FBL | FBL Entrez, Source | fibrillarin | 10535 | -0.063 | -0.4339 | Yes |
| 21 | PLAU | PLAU Entrez, Source | plasminogen activator, urokinase | 10548 | -0.063 | -0.4168 | Yes |
| 22 | VEGF | VEGF Entrez, Source | vascular endothelial growth factor | 10686 | -0.066 | -0.4083 | Yes |
| 23 | XPO7 | XPO7 Entrez, Source | exportin 7 | 11164 | -0.077 | -0.4222 | Yes |
| 24 | CTSC | CTSC Entrez, Source | cathepsin C | 11373 | -0.083 | -0.4140 | Yes |
| 25 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 11496 | -0.086 | -0.3987 | Yes |
| 26 | TM7SF2 | TM7SF2 Entrez, Source | transmembrane 7 superfamily member 2 | 11819 | -0.096 | -0.3954 | Yes |
| 27 | ALG3 | ALG3 Entrez, Source | asparagine-linked glycosylation 3 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase) | 12125 | -0.108 | -0.3875 | Yes |
| 28 | PRR3 | PRR3 Entrez, Source | proline rich 3 | 12301 | -0.116 | -0.3676 | Yes |
| 29 | PMVK | PMVK Entrez, Source | phosphomevalonate kinase | 12335 | -0.118 | -0.3364 | Yes |
| 30 | TRMT1 | TRMT1 Entrez, Source | TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae) | 12818 | -0.154 | -0.3286 | Yes |
| 31 | DDX46 | DDX46 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 | 12972 | -0.176 | -0.2899 | Yes |
| 32 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 13022 | -0.190 | -0.2395 | Yes |
| 33 | SLC29A1 | SLC29A1 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 1 | 13027 | -0.192 | -0.1850 | Yes |
| 34 | CA2 | CA2 Entrez, Source | carbonic anhydrase II | 13081 | -0.204 | -0.1309 | Yes |
| 35 | JAK2 | JAK2 Entrez, Source | Janus kinase 2 (a protein tyrosine kinase) | 13193 | -0.251 | -0.0678 | Yes |
| 36 | PSPH | PSPH Entrez, Source | phosphoserine phosphatase | 13229 | -0.277 | 0.0085 | Yes |