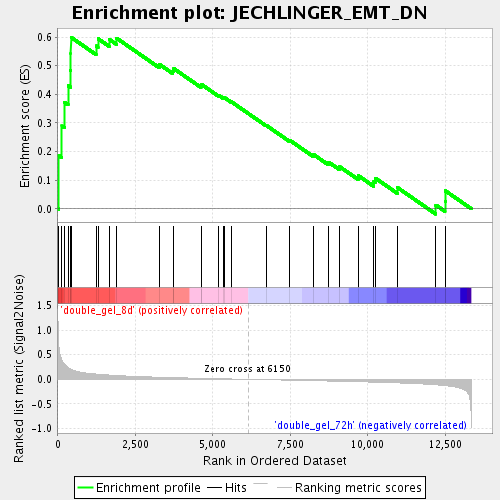
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | JECHLINGER_EMT_DN |
| Enrichment Score (ES) | 0.59970963 |
| Normalized Enrichment Score (NES) | 1.771613 |
| Nominal p-value | 0.0036429872 |
| FDR q-value | 0.03647977 |
| FWER p-Value | 0.995 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 32 | 0.660 | 0.1857 | Yes |
| 2 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 130 | 0.390 | 0.2897 | Yes |
| 3 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 218 | 0.309 | 0.3713 | Yes |
| 4 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 330 | 0.237 | 0.4306 | Yes |
| 5 | TIMP3 | TIMP3 Entrez, Source | TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 406 | 0.208 | 0.4843 | Yes |
| 6 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 418 | 0.205 | 0.5418 | Yes |
| 7 | TACSTD1 | TACSTD1 Entrez, Source | tumor-associated calcium signal transducer 1 | 422 | 0.204 | 0.5997 | Yes |
| 8 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 1235 | 0.108 | 0.5695 | No |
| 9 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 1297 | 0.104 | 0.5946 | No |
| 10 | SERPINB5 | SERPINB5 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 5 | 1661 | 0.086 | 0.5920 | No |
| 11 | VAMP8 | VAMP8 Entrez, Source | vesicle-associated membrane protein 8 (endobrevin) | 1893 | 0.077 | 0.5965 | No |
| 12 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 3274 | 0.043 | 0.5051 | No |
| 13 | ITGB5 | ITGB5 Entrez, Source | integrin, beta 5 | 3714 | 0.035 | 0.4821 | No |
| 14 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 3739 | 0.035 | 0.4901 | No |
| 15 | NUMB | NUMB Entrez, Source | numb homolog (Drosophila) | 4620 | 0.021 | 0.4299 | No |
| 16 | ATP1A1 | ATP1A1 Entrez, Source | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 4640 | 0.020 | 0.4343 | No |
| 17 | ACTN4 | ACTN4 Entrez, Source | actinin, alpha 4 | 5181 | 0.013 | 0.3974 | No |
| 18 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 5353 | 0.010 | 0.3875 | No |
| 19 | CTSH | CTSH Entrez, Source | cathepsin H | 5387 | 0.010 | 0.3878 | No |
| 20 | PADI2 | PADI2 Entrez, Source | peptidyl arginine deiminase, type II | 5592 | 0.007 | 0.3746 | No |
| 21 | STAT5A | STAT5A Entrez, Source | signal transducer and activator of transcription 5A | 6719 | -0.007 | 0.2921 | No |
| 22 | HMMR | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 7489 | -0.017 | 0.2390 | No |
| 23 | KITLG | KITLG Entrez, Source | KIT ligand | 8254 | -0.027 | 0.1894 | No |
| 24 | RARA | RARA Entrez, Source | retinoic acid receptor, alpha | 8732 | -0.034 | 0.1632 | No |
| 25 | ARHGEF1 | ARHGEF1 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 1 | 9085 | -0.039 | 0.1478 | No |
| 26 | TGM2 | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 9694 | -0.049 | 0.1159 | No |
| 27 | GRB7 | GRB7 Entrez, Source | growth factor receptor-bound protein 7 | 10194 | -0.057 | 0.0946 | No |
| 28 | JUP | JUP Entrez, Source | junction plakoglobin | 10241 | -0.057 | 0.1075 | No |
| 29 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 10957 | -0.072 | 0.0744 | No |
| 30 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 12195 | -0.111 | 0.0132 | No |
| 31 | MYH9 | MYH9 Entrez, Source | myosin, heavy chain 9, non-muscle | 12494 | -0.127 | 0.0271 | No |
| 32 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 12504 | -0.128 | 0.0630 | No |

