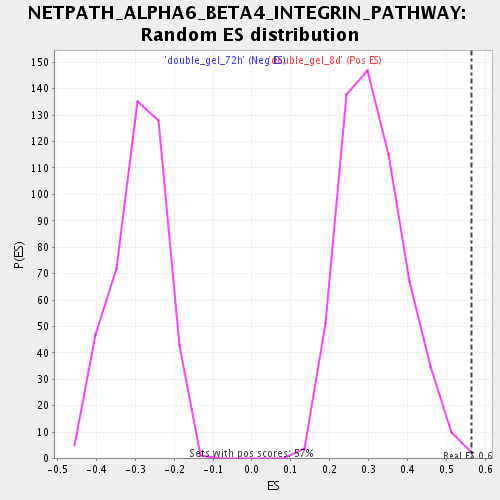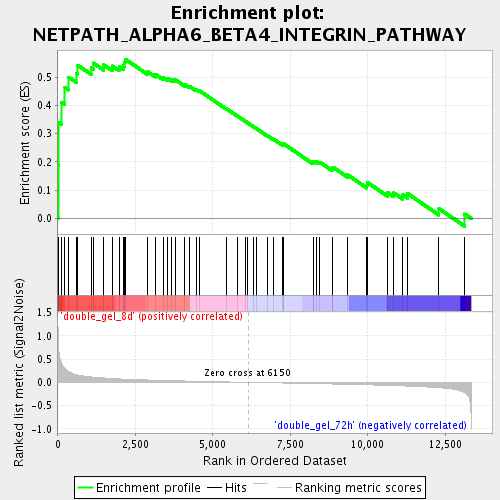
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | NETPATH_ALPHA6_BETA4_INTEGRIN_PATHWAY |
| Enrichment Score (ES) | 0.5637536 |
| Normalized Enrichment Score (NES) | 1.807936 |
| Nominal p-value | 0.0017574693 |
| FDR q-value | 0.030788044 |
| FWER p-Value | 0.965 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VIM | VIM Entrez, Source | vimentin | 6 | 0.953 | 0.1884 | Yes |
| 2 | LAMC2 | LAMC2 Entrez, Source | laminin, gamma 2 | 14 | 0.770 | 0.3404 | Yes |
| 3 | ITGB4 | ITGB4 Entrez, Source | integrin, beta 4 | 121 | 0.401 | 0.4118 | Yes |
| 4 | LAMC1 | LAMC1 Entrez, Source | laminin, gamma 1 (formerly LAMB2) | 224 | 0.303 | 0.4641 | Yes |
| 5 | LAMB2 | LAMB2 Entrez, Source | laminin, beta 2 (laminin S) | 351 | 0.229 | 0.5001 | Yes |
| 6 | AR | AR Entrez, Source | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; Kennedy disease) | 602 | 0.164 | 0.5138 | Yes |
| 7 | MMP7 | MMP7 Entrez, Source | matrix metallopeptidase 7 (matrilysin, uterine) | 641 | 0.159 | 0.5423 | Yes |
| 8 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 1072 | 0.116 | 0.5330 | Yes |
| 9 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 1140 | 0.113 | 0.5503 | Yes |
| 10 | RTKN | RTKN Entrez, Source | rhotekin | 1476 | 0.095 | 0.5439 | Yes |
| 11 | BAD | BAD Entrez, Source | BCL2-antagonist of cell death | 1750 | 0.083 | 0.5397 | Yes |
| 12 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 1975 | 0.074 | 0.5375 | Yes |
| 13 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 2118 | 0.069 | 0.5405 | Yes |
| 14 | CLCA2 | CLCA2 Entrez, Source | chloride channel, calcium activated, family member 2 | 2146 | 0.069 | 0.5521 | Yes |
| 15 | CD151 | CD151 Entrez, Source | CD151 molecule (Raph blood group) | 2171 | 0.068 | 0.5638 | Yes |
| 16 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 2895 | 0.051 | 0.5194 | No |
| 17 | RHOA | RHOA Entrez, Source | ras homolog gene family, member A | 3147 | 0.045 | 0.5095 | No |
| 18 | PRKCD | PRKCD Entrez, Source | protein kinase C, delta | 3396 | 0.041 | 0.4989 | No |
| 19 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 3538 | 0.038 | 0.4958 | No |
| 20 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 3679 | 0.036 | 0.4923 | No |
| 21 | SRC | SRC Entrez, Source | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | 3782 | 0.034 | 0.4914 | No |
| 22 | EIF4EBP1 | EIF4EBP1 Entrez, Source | eukaryotic translation initiation factor 4E binding protein 1 | 4091 | 0.029 | 0.4739 | No |
| 23 | PIK3R3 | PIK3R3 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 4251 | 0.026 | 0.4671 | No |
| 24 | PIK3CA | PIK3CA Entrez, Source | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 4457 | 0.023 | 0.4562 | No |
| 25 | CASP3 | CASP3 Entrez, Source | caspase 3, apoptosis-related cysteine peptidase | 4567 | 0.022 | 0.4523 | No |
| 26 | DSP | DSP Entrez, Source | desmoplakin | 5452 | 0.009 | 0.3876 | No |
| 27 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 5809 | 0.004 | 0.3617 | No |
| 28 | NTN1 | NTN1 Entrez, Source | netrin 1 | 6053 | 0.001 | 0.3437 | No |
| 29 | RAC1 | RAC1 Entrez, Source | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 6128 | 0.000 | 0.3382 | No |
| 30 | LAMB3 | LAMB3 Entrez, Source | laminin, beta 3 | 6323 | -0.002 | 0.3240 | No |
| 31 | LAMA3 | LAMA3 Entrez, Source | laminin, alpha 3 | 6404 | -0.003 | 0.3186 | No |
| 32 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 6769 | -0.008 | 0.2928 | No |
| 33 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 6967 | -0.010 | 0.2800 | No |
| 34 | LAMA5 | LAMA5 Entrez, Source | laminin, alpha 5 | 7254 | -0.014 | 0.2612 | No |
| 35 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 7269 | -0.014 | 0.2629 | No |
| 36 | ITGB4BP | ITGB4BP Entrez, Source | integrin beta 4 binding protein | 7280 | -0.014 | 0.2649 | No |
| 37 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 8235 | -0.027 | 0.1984 | No |
| 38 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8261 | -0.027 | 0.2020 | No |
| 39 | YWHAZ | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 8338 | -0.028 | 0.2019 | No |
| 40 | IRS1 | IRS1 Entrez, Source | insulin receptor substrate 1 | 8456 | -0.030 | 0.1990 | No |
| 41 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 8847 | -0.035 | 0.1767 | No |
| 42 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 8872 | -0.036 | 0.1820 | No |
| 43 | METAP2 | METAP2 Entrez, Source | methionyl aminopeptidase 2 | 9353 | -0.043 | 0.1543 | No |
| 44 | RPSA | RPSA Entrez, Source | ribosomal protein SA | 9973 | -0.053 | 0.1183 | No |
| 45 | PIK3R2 | PIK3R2 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 2 (p85 beta) | 9995 | -0.053 | 0.1273 | No |
| 46 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 10639 | -0.065 | 0.0918 | No |
| 47 | AKT1 | AKT1 Entrez, Source | v-akt murine thymoma viral oncogene homolog 1 | 10838 | -0.069 | 0.0906 | No |
| 48 | GRB2 | GRB2 Entrez, Source | growth factor receptor-bound protein 2 | 11132 | -0.077 | 0.0837 | No |
| 49 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 11286 | -0.081 | 0.0883 | No |
| 50 | PLEC1 | PLEC1 Entrez, Source | plectin 1, intermediate filament binding protein 500kDa | 12297 | -0.116 | 0.0352 | No |
| 51 | FRAP1 | FRAP1 Entrez, Source | FK506 binding protein 12-rapamycin associated protein 1 | 13129 | -0.219 | 0.0160 | No |