Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | PENG_GLUTAMINE_DN |
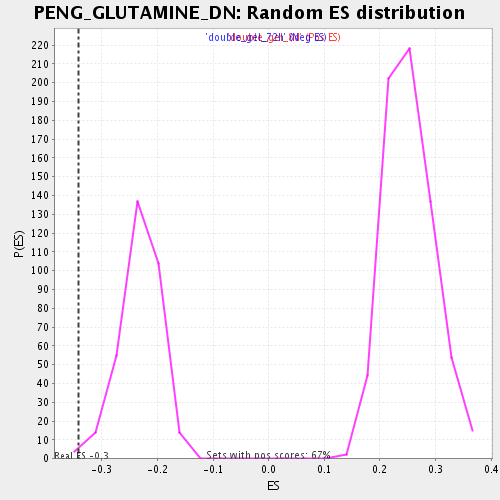
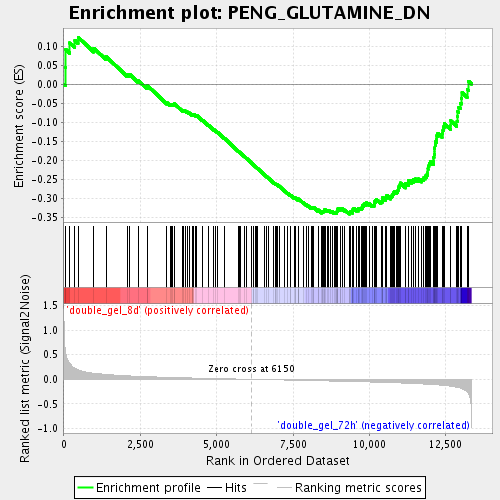
| Enrichment Score (ES) | -0.3412354 |
| Normalized Enrichment Score (NES) | -1.4688557 |
| Nominal p-value | 0.0060975607 |
| FDR q-value | 0.37899005 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 52 | 0.576 | 0.0446 | No |
| 2 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 61 | 0.555 | 0.0907 | No |
| 3 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 192 | 0.328 | 0.1084 | No |
| 4 | EMP3 | EMP3 Entrez, Source | epithelial membrane protein 3 | 358 | 0.227 | 0.1149 | No |
| 5 | CLIC1 | CLIC1 Entrez, Source | chloride intracellular channel 1 | 463 | 0.192 | 0.1232 | No |
| 6 | KHSRP | KHSRP Entrez, Source | KH-type splicing regulatory protein (FUSE binding protein 2) | 977 | 0.123 | 0.0946 | No |
| 7 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 1380 | 0.100 | 0.0725 | No |
| 8 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 2079 | 0.071 | 0.0254 | No |
| 9 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 2140 | 0.069 | 0.0266 | No |
| 10 | PSMD8 | PSMD8 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 2441 | 0.061 | 0.0090 | No |
| 11 | SLC19A1 | SLC19A1 Entrez, Source | solute carrier family 19 (folate transporter), member 1 | 2726 | 0.054 | -0.0080 | No |
| 12 | PSMA6 | PSMA6 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 6 | 2734 | 0.054 | -0.0040 | No |
| 13 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 3372 | 0.041 | -0.0489 | No |
| 14 | MYD88 | MYD88 Entrez, Source | myeloid differentiation primary response gene (88) | 3471 | 0.039 | -0.0531 | No |
| 15 | IDH3A | IDH3A Entrez, Source | isocitrate dehydrogenase 3 (NAD+) alpha | 3505 | 0.039 | -0.0523 | No |
| 16 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 3568 | 0.037 | -0.0539 | No |
| 17 | SYNGR2 | SYNGR2 Entrez, Source | synaptogyrin 2 | 3612 | 0.037 | -0.0541 | No |
| 18 | SF3B4 | SF3B4 Entrez, Source | splicing factor 3b, subunit 4, 49kDa | 3618 | 0.037 | -0.0514 | No |
| 19 | CELSR2 | CELSR2 Entrez, Source | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | 3880 | 0.032 | -0.0685 | No |
| 20 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 3915 | 0.032 | -0.0684 | No |
| 21 | TXNIP | TXNIP Entrez, Source | thioredoxin interacting protein | 3975 | 0.031 | -0.0703 | No |
| 22 | PSMD1 | PSMD1 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 4043 | 0.030 | -0.0729 | No |
| 23 | KPNA2 | KPNA2 Entrez, Source | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 4109 | 0.029 | -0.0755 | No |
| 24 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 4212 | 0.027 | -0.0810 | No |
| 25 | NUP98 | NUP98 Entrez, Source | nucleoporin 98kDa | 4217 | 0.027 | -0.0790 | No |
| 26 | PIK3R3 | PIK3R3 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 4251 | 0.026 | -0.0793 | No |
| 27 | EIF5 | EIF5 Entrez, Source | eukaryotic translation initiation factor 5 | 4315 | 0.025 | -0.0820 | No |
| 28 | BCAP31 | BCAP31 Entrez, Source | B-cell receptor-associated protein 31 | 4339 | 0.025 | -0.0816 | No |
| 29 | HSPA8 | HSPA8 Entrez, Source | heat shock 70kDa protein 8 | 4537 | 0.022 | -0.0948 | No |
| 30 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 4727 | 0.019 | -0.1075 | No |
| 31 | NMT1 | NMT1 Entrez, Source | N-myristoyltransferase 1 | 4905 | 0.017 | -0.1195 | No |
| 32 | PPP2R2A | PPP2R2A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | 4968 | 0.016 | -0.1229 | No |
| 33 | SCD | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 5035 | 0.015 | -0.1267 | No |
| 34 | PGAM1 | PGAM1 Entrez, Source | phosphoglycerate mutase 1 (brain) | 5246 | 0.012 | -0.1416 | No |
| 35 | PPIB | PPIB Entrez, Source | peptidylprolyl isomerase B (cyclophilin B) | 5713 | 0.006 | -0.1766 | No |
| 36 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 5742 | 0.005 | -0.1783 | No |
| 37 | PSMD12 | PSMD12 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 5781 | 0.005 | -0.1808 | No |
| 38 | PPIF | PPIF Entrez, Source | peptidylprolyl isomerase F (cyclophilin F) | 5913 | 0.003 | -0.1905 | No |
| 39 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 5974 | 0.002 | -0.1948 | No |
| 40 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 6135 | 0.000 | -0.2069 | No |
| 41 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 6189 | -0.000 | -0.2109 | No |
| 42 | IFI30 | IFI30 Entrez, Source | interferon, gamma-inducible protein 30 | 6191 | -0.000 | -0.2110 | No |
| 43 | CCT6A | CCT6A Entrez, Source | chaperonin containing TCP1, subunit 6A (zeta 1) | 6256 | -0.001 | -0.2157 | No |
| 44 | ATP1B3 | ATP1B3 Entrez, Source | ATPase, Na+/K+ transporting, beta 3 polypeptide | 6301 | -0.002 | -0.2189 | No |
| 45 | LAMB3 | LAMB3 Entrez, Source | laminin, beta 3 | 6323 | -0.002 | -0.2203 | No |
| 46 | HSPA5 | HSPA5 Entrez, Source | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 6557 | -0.005 | -0.2376 | No |
| 47 | DNAJB6 | DNAJB6 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 6 | 6644 | -0.006 | -0.2436 | No |
| 48 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 6700 | -0.007 | -0.2472 | No |
| 49 | PRDX1 | PRDX1 Entrez, Source | peroxiredoxin 1 | 6870 | -0.009 | -0.2592 | No |
| 50 | PSMB3 | PSMB3 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 3 | 6921 | -0.010 | -0.2622 | No |
| 51 | TCEB1 | TCEB1 Entrez, Source | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | 6931 | -0.010 | -0.2621 | No |
| 52 | ADRM1 | ADRM1 Entrez, Source | adhesion regulating molecule 1 | 6953 | -0.010 | -0.2628 | No |
| 53 | OGFR | OGFR Entrez, Source | opioid growth factor receptor | 6989 | -0.011 | -0.2646 | No |
| 54 | ADAM19 | ADAM19 Entrez, Source | ADAM metallopeptidase domain 19 (meltrin beta) | 7040 | -0.011 | -0.2674 | No |
| 55 | PFN1 | PFN1 Entrez, Source | profilin 1 | 7229 | -0.013 | -0.2806 | No |
| 56 | PTMS | PTMS Entrez, Source | parathymosin | 7316 | -0.014 | -0.2859 | No |
| 57 | LMNB1 | LMNB1 Entrez, Source | lamin B1 | 7411 | -0.015 | -0.2917 | No |
| 58 | VCP | VCP Entrez, Source | valosin-containing protein | 7423 | -0.016 | -0.2913 | No |
| 59 | GRIN2A | GRIN2A Entrez, Source | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 7544 | -0.017 | -0.2989 | No |
| 60 | HRAS | HRAS Entrez, Source | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | 7568 | -0.018 | -0.2992 | No |
| 61 | MORF4L2 | MORF4L2 Entrez, Source | mortality factor 4 like 2 | 7580 | -0.018 | -0.2985 | No |
| 62 | PSMD3 | PSMD3 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | 7666 | -0.019 | -0.3034 | No |
| 63 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 7672 | -0.019 | -0.3021 | No |
| 64 | DNAJA1 | DNAJA1 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 1 | 7683 | -0.019 | -0.3013 | No |
| 65 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 7828 | -0.021 | -0.3104 | No |
| 66 | COPS3 | COPS3 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | 7930 | -0.022 | -0.3162 | No |
| 67 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 8004 | -0.024 | -0.3198 | No |
| 68 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 8111 | -0.025 | -0.3257 | No |
| 69 | NDUFS6 | NDUFS6 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) | 8128 | -0.025 | -0.3248 | No |
| 70 | CCT2 | CCT2 Entrez, Source | chaperonin containing TCP1, subunit 2 (beta) | 8167 | -0.026 | -0.3255 | No |
| 71 | RANGAP1 | RANGAP1 Entrez, Source | Ran GTPase activating protein 1 | 8169 | -0.026 | -0.3234 | No |
| 72 | PHB | PHB Entrez, Source | prohibitin | 8324 | -0.028 | -0.3327 | No |
| 73 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 8325 | -0.028 | -0.3303 | No |
| 74 | BMS1L | BMS1L Entrez, Source | BMS1-like, ribosome assembly protein (yeast) | 8437 | -0.030 | -0.3362 | No |
| 75 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 8447 | -0.030 | -0.3344 | No |
| 76 | RNASEH1 | RNASEH1 Entrez, Source | ribonuclease H1 | 8474 | -0.031 | -0.3338 | No |
| 77 | TCP1 | TCP1 Entrez, Source | t-complex 1 | 8504 | -0.031 | -0.3334 | No |
| 78 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 8524 | -0.031 | -0.3322 | No |
| 79 | ADCY3 | ADCY3 Entrez, Source | adenylate cyclase 3 | 8531 | -0.031 | -0.3301 | No |
| 80 | EIF2B4 | EIF2B4 Entrez, Source | eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa | 8566 | -0.032 | -0.3300 | No |
| 81 | BZW1 | BZW1 Entrez, Source | basic leucine zipper and W2 domains 1 | 8626 | -0.033 | -0.3317 | No |
| 82 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 8663 | -0.033 | -0.3316 | No |
| 83 | CALR | CALR Entrez, Source | calreticulin | 8727 | -0.034 | -0.3336 | No |
| 84 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 8780 | -0.035 | -0.3346 | No |
| 85 | RBBP6 | RBBP6 Entrez, Source | retinoblastoma binding protein 6 | 8839 | -0.035 | -0.3360 | No |
| 86 | BOP1 | BOP1 Entrez, Source | block of proliferation 1 | 8881 | -0.036 | -0.3361 | No |
| 87 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 8929 | -0.037 | -0.3366 | No |
| 88 | POLR2C | POLR2C Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 8936 | -0.037 | -0.3340 | No |
| 89 | EIF5A | EIF5A Entrez, Source | eukaryotic translation initiation factor 5A | 8947 | -0.037 | -0.3316 | No |
| 90 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 8961 | -0.037 | -0.3295 | No |
| 91 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 8969 | -0.037 | -0.3269 | No |
| 92 | NUDC | NUDC Entrez, Source | nuclear distribution gene C homolog (A. nidulans) | 9037 | -0.038 | -0.3288 | No |
| 93 | PABPN1 | PABPN1 Entrez, Source | poly(A) binding protein, nuclear 1 | 9064 | -0.038 | -0.3275 | No |
| 94 | HNRPAB | HNRPAB Entrez, Source | heterogeneous nuclear ribonucleoprotein A/B | 9101 | -0.039 | -0.3270 | No |
| 95 | PPP1R10 | PPP1R10 Entrez, Source | protein phosphatase 1, regulatory subunit 10 | 9190 | -0.040 | -0.3303 | No |
| 96 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 9335 | -0.042 | -0.3377 | Yes |
| 97 | SSBP1 | SSBP1 Entrez, Source | single-stranded DNA binding protein 1 | 9358 | -0.043 | -0.3358 | Yes |
| 98 | DKC1 | DKC1 Entrez, Source | dyskeratosis congenita 1, dyskerin | 9386 | -0.043 | -0.3342 | Yes |
| 99 | NCL | NCL Entrez, Source | nucleolin | 9448 | -0.044 | -0.3351 | Yes |
| 100 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 9455 | -0.044 | -0.3318 | Yes |
| 101 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 9457 | -0.044 | -0.3282 | Yes |
| 102 | CCT5 | CCT5 Entrez, Source | chaperonin containing TCP1, subunit 5 (epsilon) | 9486 | -0.045 | -0.3265 | Yes |
| 103 | PRPF4 | PRPF4 Entrez, Source | PRP4 pre-mRNA processing factor 4 homolog (yeast) | 9579 | -0.046 | -0.3296 | Yes |
| 104 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 9628 | -0.047 | -0.3292 | Yes |
| 105 | SQLE | SQLE Entrez, Source | squalene epoxidase | 9644 | -0.048 | -0.3264 | Yes |
| 106 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 9687 | -0.048 | -0.3255 | Yes |
| 107 | ABCF1 | ABCF1 Entrez, Source | ATP-binding cassette, sub-family F (GCN20), member 1 | 9738 | -0.049 | -0.3251 | Yes |
| 108 | PRODH | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 9764 | -0.050 | -0.3229 | Yes |
| 109 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 9770 | -0.050 | -0.3190 | Yes |
| 110 | G3BP | G3BP Entrez, Source | - | 9801 | -0.050 | -0.3171 | Yes |
| 111 | RQCD1 | RQCD1 Entrez, Source | RCD1 required for cell differentiation1 homolog (S. pombe) | 9821 | -0.051 | -0.3143 | Yes |
| 112 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 9884 | -0.052 | -0.3146 | Yes |
| 113 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 9899 | -0.052 | -0.3113 | Yes |
| 114 | PIK3R2 | PIK3R2 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 2 (p85 beta) | 9995 | -0.053 | -0.3141 | Yes |
| 115 | BYSL | BYSL Entrez, Source | bystin-like | 10106 | -0.055 | -0.3178 | Yes |
| 116 | PTDSS1 | PTDSS1 Entrez, Source | phosphatidylserine synthase 1 | 10150 | -0.056 | -0.3163 | Yes |
| 117 | ALG8 | ALG8 Entrez, Source | asparagine-linked glycosylation 8 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | 10169 | -0.056 | -0.3130 | Yes |
| 118 | GMPS | GMPS Entrez, Source | guanine monphosphate synthetase | 10172 | -0.056 | -0.3084 | Yes |
| 119 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 10199 | -0.057 | -0.3056 | Yes |
| 120 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 10227 | -0.057 | -0.3028 | Yes |
| 121 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 10378 | -0.060 | -0.3092 | Yes |
| 122 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 10415 | -0.061 | -0.3068 | Yes |
| 123 | EWSR1 | EWSR1 Entrez, Source | Ewing sarcoma breakpoint region 1 | 10419 | -0.061 | -0.3019 | Yes |
| 124 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 10434 | -0.061 | -0.2979 | Yes |
| 125 | FBL | FBL Entrez, Source | fibrillarin | 10535 | -0.063 | -0.3002 | Yes |
| 126 | PSMB6 | PSMB6 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 6 | 10550 | -0.063 | -0.2959 | Yes |
| 127 | HNRPL | HNRPL Entrez, Source | heterogeneous nuclear ribonucleoprotein L | 10566 | -0.064 | -0.2917 | Yes |
| 128 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 10700 | -0.066 | -0.2962 | Yes |
| 129 | RPS26 | RPS26 Entrez, Source | ribosomal protein S26 | 10735 | -0.067 | -0.2932 | Yes |
| 130 | SGTA | SGTA Entrez, Source | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | 10760 | -0.067 | -0.2893 | Yes |
| 131 | TBL3 | TBL3 Entrez, Source | transducin (beta)-like 3 | 10783 | -0.068 | -0.2853 | Yes |
| 132 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 10803 | -0.068 | -0.2809 | Yes |
| 133 | SRPK1 | SRPK1 Entrez, Source | SFRS protein kinase 1 | 10895 | -0.071 | -0.2819 | Yes |
| 134 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 10929 | -0.072 | -0.2784 | Yes |
| 135 | RNF126 | RNF126 Entrez, Source | ring finger protein 126 | 10941 | -0.072 | -0.2732 | Yes |
| 136 | PDCD4 | PDCD4 Entrez, Source | programmed cell death 4 (neoplastic transformation inhibitor) | 10963 | -0.073 | -0.2687 | Yes |
| 137 | SSB | SSB Entrez, Source | Sjogren syndrome antigen B (autoantigen La) | 10994 | -0.073 | -0.2648 | Yes |
| 138 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 11005 | -0.073 | -0.2594 | Yes |
| 139 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 11182 | -0.078 | -0.2662 | Yes |
| 140 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11187 | -0.078 | -0.2599 | Yes |
| 141 | MRPL23 | MRPL23 Entrez, Source | mitochondrial ribosomal protein L23 | 11272 | -0.081 | -0.2595 | Yes |
| 142 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 11282 | -0.081 | -0.2534 | Yes |
| 143 | CTSC | CTSC Entrez, Source | cathepsin C | 11373 | -0.083 | -0.2532 | Yes |
| 144 | DDX18 | DDX18 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 | 11433 | -0.085 | -0.2506 | Yes |
| 145 | C1QBP | C1QBP Entrez, Source | complement component 1, q subcomponent binding protein | 11506 | -0.086 | -0.2488 | Yes |
| 146 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 11605 | -0.089 | -0.2487 | Yes |
| 147 | FDFT1 | FDFT1 Entrez, Source | farnesyl-diphosphate farnesyltransferase 1 | 11716 | -0.093 | -0.2493 | Yes |
| 148 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 11781 | -0.095 | -0.2462 | Yes |
| 149 | NOL5A | NOL5A Entrez, Source | nucleolar protein 5A (56kDa with KKE/D repeat) | 11821 | -0.096 | -0.2410 | Yes |
| 150 | DDX1 | DDX1 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 11880 | -0.098 | -0.2371 | Yes |
| 151 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 11909 | -0.100 | -0.2309 | Yes |
| 152 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 11914 | -0.100 | -0.2228 | Yes |
| 153 | TAP1 | TAP1 Entrez, Source | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 11920 | -0.100 | -0.2148 | Yes |
| 154 | EBP | EBP Entrez, Source | emopamil binding protein (sterol isomerase) | 11949 | -0.101 | -0.2084 | Yes |
| 155 | PTPN6 | PTPN6 Entrez, Source | protein tyrosine phosphatase, non-receptor type 6 | 12001 | -0.103 | -0.2036 | Yes |
| 156 | PSME4 | PSME4 Entrez, Source | proteasome (prosome, macropain) activator subunit 4 | 12096 | -0.107 | -0.2018 | Yes |
| 157 | SLC29A2 | SLC29A2 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 2 | 12103 | -0.107 | -0.1932 | Yes |
| 158 | SLC5A6 | SLC5A6 Entrez, Source | solute carrier family 5 (sodium-dependent vitamin transporter), member 6 | 12113 | -0.108 | -0.1848 | Yes |
| 159 | ALG3 | ALG3 Entrez, Source | asparagine-linked glycosylation 3 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase) | 12125 | -0.108 | -0.1765 | Yes |
| 160 | EIF2B5 | EIF2B5 Entrez, Source | eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa | 12127 | -0.108 | -0.1674 | Yes |
| 161 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 12144 | -0.109 | -0.1595 | Yes |
| 162 | IDI1 | IDI1 Entrez, Source | isopentenyl-diphosphate delta isomerase 1 | 12159 | -0.110 | -0.1513 | Yes |
| 163 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 12195 | -0.111 | -0.1446 | Yes |
| 164 | FUS | FUS Entrez, Source | fusion (involved in t(12;16) in malignant liposarcoma) | 12205 | -0.112 | -0.1359 | Yes |
| 165 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 12235 | -0.113 | -0.1286 | Yes |
| 166 | FKBP4 | FKBP4 Entrez, Source | FK506 binding protein 4, 59kDa | 12376 | -0.120 | -0.1291 | Yes |
| 167 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 12402 | -0.121 | -0.1208 | Yes |
| 168 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 12425 | -0.123 | -0.1121 | Yes |
| 169 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 12447 | -0.123 | -0.1033 | Yes |
| 170 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 12656 | -0.138 | -0.1075 | Yes |
| 171 | XBP1 | XBP1 Entrez, Source | X-box binding protein 1 | 12661 | -0.138 | -0.0962 | Yes |
| 172 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 12862 | -0.160 | -0.0979 | Yes |
| 173 | TDG | TDG Entrez, Source | thymine-DNA glycosylase | 12879 | -0.162 | -0.0855 | Yes |
| 174 | INHBC | INHBC Entrez, Source | inhibin, beta C | 12890 | -0.163 | -0.0725 | Yes |
| 175 | SRM | SRM Entrez, Source | spermidine synthase | 12923 | -0.168 | -0.0608 | Yes |
| 176 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 12976 | -0.177 | -0.0498 | Yes |
| 177 | LIMK2 | LIMK2 Entrez, Source | LIM domain kinase 2 | 13008 | -0.187 | -0.0365 | Yes |
| 178 | SLC29A1 | SLC29A1 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 1 | 13027 | -0.192 | -0.0216 | Yes |
| 179 | SFPQ | SFPQ Entrez, Source | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | 13196 | -0.252 | -0.0132 | Yes |
| 180 | FASN | FASN Entrez, Source | fatty acid synthase | 13241 | -0.288 | 0.0077 | Yes |