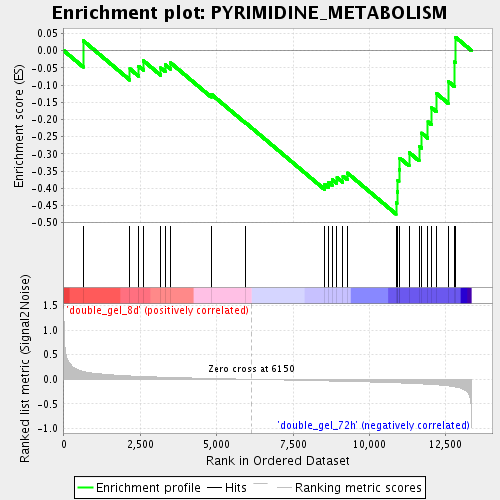
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.4749834 |
| Normalized Enrichment Score (NES) | -1.4325228 |
| Nominal p-value | 0.06521739 |
| FDR q-value | 0.42968643 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DPYD | DPYD Entrez, Source | dihydropyrimidine dehydrogenase | 633 | 0.160 | 0.0292 | No |
| 2 | NUDT2 | NUDT2 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 2 | 2160 | 0.068 | -0.0526 | No |
| 3 | CTPS2 | CTPS2 Entrez, Source | CTP synthase II | 2446 | 0.061 | -0.0448 | No |
| 4 | POLR2G | POLR2G Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide G | 2604 | 0.057 | -0.0291 | No |
| 5 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 3170 | 0.045 | -0.0498 | No |
| 6 | CMPK | CMPK Entrez, Source | cytidylate kinase | 3318 | 0.042 | -0.0407 | No |
| 7 | UPP1 | UPP1 Entrez, Source | uridine phosphorylase 1 | 3489 | 0.039 | -0.0349 | No |
| 8 | AK3 | AK3 Entrez, Source | adenylate kinase 3 | 4820 | 0.018 | -0.1262 | No |
| 9 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 5958 | 0.003 | -0.2103 | No |
| 10 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 8535 | -0.031 | -0.3888 | No |
| 11 | DHODH | DHODH Entrez, Source | dihydroorotate dehydrogenase | 8664 | -0.033 | -0.3825 | No |
| 12 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 8780 | -0.035 | -0.3746 | No |
| 13 | POLR2C | POLR2C Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 8936 | -0.037 | -0.3686 | No |
| 14 | NME2 | NME2 Entrez, Source | non-metastatic cells 2, protein (NM23B) expressed in | 9131 | -0.039 | -0.3644 | No |
| 15 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 9276 | -0.041 | -0.3553 | No |
| 16 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 10870 | -0.070 | -0.4413 | Yes |
| 17 | CANT1 | CANT1 Entrez, Source | calcium activated nucleotidase 1 | 10903 | -0.071 | -0.4098 | Yes |
| 18 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 10927 | -0.072 | -0.3771 | Yes |
| 19 | ENTPD1 | ENTPD1 Entrez, Source | ectonucleoside triphosphate diphosphohydrolase 1 | 10967 | -0.073 | -0.3452 | Yes |
| 20 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 10993 | -0.073 | -0.3120 | Yes |
| 21 | NT5E | NT5E Entrez, Source | 5'-nucleotidase, ecto (CD73) | 11299 | -0.081 | -0.2959 | Yes |
| 22 | POLB | POLB Entrez, Source | polymerase (DNA directed), beta | 11626 | -0.090 | -0.2774 | Yes |
| 23 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 11707 | -0.092 | -0.2392 | Yes |
| 24 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 11914 | -0.100 | -0.2068 | Yes |
| 25 | UPB1 | UPB1 Entrez, Source | ureidopropionase, beta | 12035 | -0.104 | -0.1658 | Yes |
| 26 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 12199 | -0.111 | -0.1246 | Yes |
| 27 | POLR1B | POLR1B Entrez, Source | polymerase (RNA) I polypeptide B, 128kDa | 12576 | -0.133 | -0.0890 | Yes |
| 28 | DCK | DCK Entrez, Source | deoxycytidine kinase | 12784 | -0.151 | -0.0323 | Yes |
| 29 | AK3L1 | AK3L1 Entrez, Source | adenylate kinase 3-like 1 | 12819 | -0.155 | 0.0393 | Yes |

