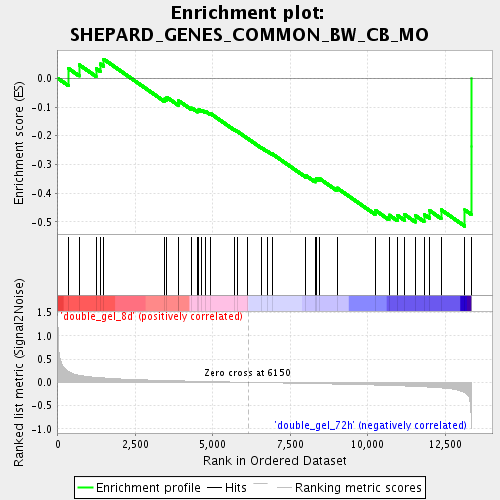
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | SHEPARD_GENES_COMMON_BW_CB_MO |
| Enrichment Score (ES) | -0.51525784 |
| Normalized Enrichment Score (NES) | -1.6657649 |
| Nominal p-value | 0.009029346 |
| FDR q-value | 0.18779358 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UCP2 | UCP2 Entrez, Source | uncoupling protein 2 (mitochondrial, proton carrier) | 344 | 0.231 | 0.0340 | No |
| 2 | POU3F1 | POU3F1 Entrez, Source | POU domain, class 3, transcription factor 1 | 689 | 0.151 | 0.0472 | No |
| 3 | NR2F1 | NR2F1 Entrez, Source | nuclear receptor subfamily 2, group F, member 1 | 1234 | 0.108 | 0.0343 | No |
| 4 | SFRP1 | SFRP1 Entrez, Source | secreted frizzled-related protein 1 | 1367 | 0.100 | 0.0503 | No |
| 5 | RTKN | RTKN Entrez, Source | rhotekin | 1476 | 0.095 | 0.0669 | No |
| 6 | GOLGB1 | GOLGB1 Entrez, Source | golgi autoantigen, golgin subfamily b, macrogolgin (with transmembrane signal), 1 | 3455 | 0.040 | -0.0716 | No |
| 7 | POU3F2 | POU3F2 Entrez, Source | POU domain, class 3, transcription factor 2 | 3514 | 0.038 | -0.0660 | No |
| 8 | CTSL | CTSL Entrez, Source | cathepsin L | 3892 | 0.032 | -0.0860 | No |
| 9 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 3898 | 0.032 | -0.0782 | No |
| 10 | SOX11 | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 4318 | 0.025 | -0.1031 | No |
| 11 | POU3F3 | POU3F3 Entrez, Source | POU domain, class 3, transcription factor 3 | 4509 | 0.022 | -0.1116 | No |
| 12 | TACC3 | TACC3 Entrez, Source | transforming, acidic coiled-coil containing protein 3 | 4550 | 0.022 | -0.1090 | No |
| 13 | ATP1A1 | ATP1A1 Entrez, Source | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 4640 | 0.020 | -0.1104 | No |
| 14 | MYH8 | MYH8 Entrez, Source | myosin, heavy chain 8, skeletal muscle, perinatal | 4751 | 0.019 | -0.1138 | No |
| 15 | GPSM1 | GPSM1 Entrez, Source | G-protein signalling modulator 1 (AGS3-like, C. elegans) | 4927 | 0.016 | -0.1227 | No |
| 16 | CDCA2 | CDCA2 Entrez, Source | cell division cycle associated 2 | 5699 | 0.006 | -0.1792 | No |
| 17 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 5787 | 0.005 | -0.1846 | No |
| 18 | LHX1 | LHX1 Entrez, Source | LIM homeobox 1 | 6116 | 0.000 | -0.2091 | No |
| 19 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 6574 | -0.005 | -0.2420 | No |
| 20 | FOXM1 | FOXM1 Entrez, Source | forkhead box M1 | 6751 | -0.008 | -0.2533 | No |
| 21 | TTN | TTN Entrez, Source | titin | 6911 | -0.010 | -0.2628 | No |
| 22 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 8004 | -0.024 | -0.3387 | No |
| 23 | HEY1 | HEY1 Entrez, Source | hairy/enhancer-of-split related with YRPW motif 1 | 8303 | -0.028 | -0.3539 | No |
| 24 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 8334 | -0.028 | -0.3489 | No |
| 25 | BMS1L | BMS1L Entrez, Source | BMS1-like, ribosome assembly protein (yeast) | 8437 | -0.030 | -0.3488 | No |
| 26 | OTP | OTP Entrez, Source | orthopedia homolog (Drosophila) | 9009 | -0.038 | -0.3819 | No |
| 27 | GRIK3 | GRIK3 Entrez, Source | glutamate receptor, ionotropic, kainate 3 | 10240 | -0.057 | -0.4595 | No |
| 28 | DDX17 | DDX17 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | 10688 | -0.066 | -0.4760 | No |
| 29 | MDK | MDK Entrez, Source | midkine (neurite growth-promoting factor 2) | 10954 | -0.072 | -0.4772 | No |
| 30 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11187 | -0.078 | -0.4745 | No |
| 31 | PTPRN | PTPRN Entrez, Source | protein tyrosine phosphatase, receptor type, N | 11537 | -0.088 | -0.4780 | Yes |
| 32 | TM7SF2 | TM7SF2 Entrez, Source | transmembrane 7 superfamily member 2 | 11819 | -0.096 | -0.4742 | Yes |
| 33 | AURKB | AURKB Entrez, Source | aurora kinase B | 11998 | -0.103 | -0.4610 | Yes |
| 34 | FGFR4 | FGFR4 Entrez, Source | fibroblast growth factor receptor 4 | 12369 | -0.119 | -0.4579 | Yes |
| 35 | NOTCH2 | NOTCH2 Entrez, Source | Notch homolog 2 (Drosophila) | 13133 | -0.220 | -0.4583 | Yes |
| 36 | ASCL1 | ASCL1 Entrez, Source | achaete-scute complex-like 1 (Drosophila) | 13340 | -0.915 | -0.2369 | Yes |
| 37 | EDG1 | EDG1 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | 13341 | -0.915 | 0.0001 | Yes |

