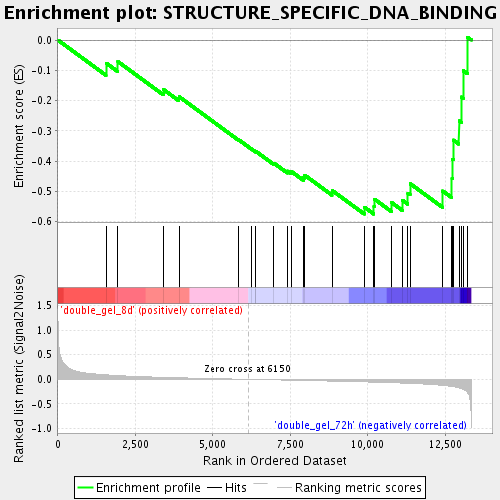
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | STRUCTURE_SPECIFIC_DNA_BINDING |
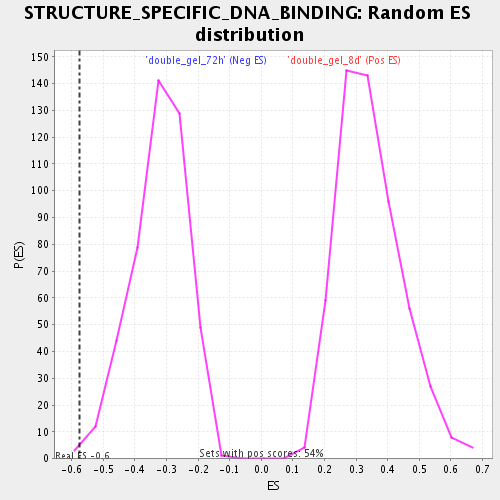
| Enrichment Score (ES) | -0.5752266 |
| Normalized Enrichment Score (NES) | -1.7748128 |
| Nominal p-value | 0.0043668123 |
| FDR q-value | 0.14844257 |
| FWER p-Value | 0.959 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TERF2IP | TERF2IP Entrez, Source | telomeric repeat binding factor 2, interacting protein | 1554 | 0.091 | -0.0760 | No |
| 2 | RBMS1 | RBMS1 Entrez, Source | RNA binding motif, single stranded interacting protein 1 | 1934 | 0.075 | -0.0710 | No |
| 3 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 3405 | 0.040 | -0.1634 | No |
| 4 | RAD23A | RAD23A Entrez, Source | RAD23 homolog A (S. cerevisiae) | 3908 | 0.032 | -0.1869 | No |
| 5 | HNRPDL | HNRPDL Entrez, Source | heterogeneous nuclear ribonucleoprotein D-like | 5830 | 0.004 | -0.3293 | No |
| 6 | WBP11 | WBP11 Entrez, Source | WW domain binding protein 11 | 6251 | -0.001 | -0.3603 | No |
| 7 | NR0B1 | NR0B1 Entrez, Source | nuclear receptor subfamily 0, group B, member 1 | 6385 | -0.003 | -0.3690 | No |
| 8 | SATB1 | SATB1 Entrez, Source | special AT-rich sequence binding protein 1 (binds to nuclear matrix/scaffold-associating DNA's) | 6392 | -0.003 | -0.3681 | No |
| 9 | FOXC2 | FOXC2 Entrez, Source | forkhead box C2 (MFH-1, mesenchyme forkhead 1) | 6970 | -0.010 | -0.4068 | No |
| 10 | MTERF | MTERF Entrez, Source | mitochondrial transcription termination factor | 7408 | -0.015 | -0.4327 | No |
| 11 | SAFB | SAFB Entrez, Source | scaffold attachment factor B | 7539 | -0.017 | -0.4348 | No |
| 12 | SUB1 | SUB1 Entrez, Source | SUB1 homolog (S. cerevisiae) | 7922 | -0.022 | -0.4536 | No |
| 13 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 7961 | -0.023 | -0.4462 | No |
| 14 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 8847 | -0.035 | -0.4969 | No |
| 15 | HLF | HLF Entrez, Source | hepatic leukemia factor | 9889 | -0.052 | -0.5520 | Yes |
| 16 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 10199 | -0.057 | -0.5500 | Yes |
| 17 | IGHMBP2 | IGHMBP2 Entrez, Source | immunoglobulin mu binding protein 2 | 10212 | -0.057 | -0.5255 | Yes |
| 18 | TREX1 | TREX1 Entrez, Source | three prime repair exonuclease 1 | 10767 | -0.068 | -0.5369 | Yes |
| 19 | PNKP | PNKP Entrez, Source | polynucleotide kinase 3'-phosphatase | 11117 | -0.076 | -0.5292 | Yes |
| 20 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 11295 | -0.081 | -0.5062 | Yes |
| 21 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 11368 | -0.083 | -0.4745 | Yes |
| 22 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 12418 | -0.122 | -0.4988 | Yes |
| 23 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 12718 | -0.143 | -0.4574 | Yes |
| 24 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 12731 | -0.144 | -0.3940 | Yes |
| 25 | MSH3 | MSH3 Entrez, Source | mutS homolog 3 (E. coli) | 12763 | -0.148 | -0.3303 | Yes |
| 26 | TERF1 | TERF1 Entrez, Source | telomeric repeat binding factor (NIMA-interacting) 1 | 12945 | -0.172 | -0.2670 | Yes |
| 27 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 13022 | -0.190 | -0.1881 | Yes |
| 28 | FUBP1 | FUBP1 Entrez, Source | far upstream element (FUSE) binding protein 1 | 13083 | -0.205 | -0.1012 | Yes |
| 29 | APTX | APTX Entrez, Source | aprataxin | 13219 | -0.270 | 0.0092 | Yes |