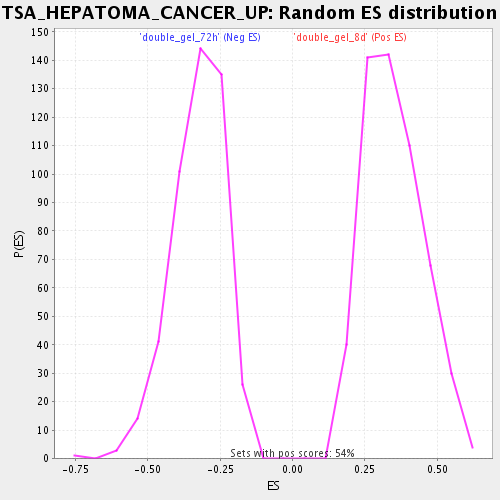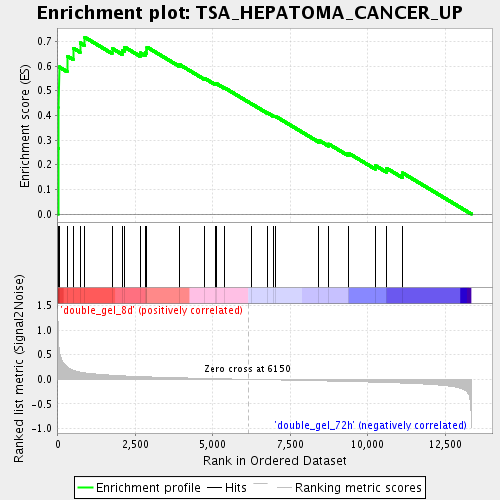
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | TSA_HEPATOMA_CANCER_UP |
| Enrichment Score (ES) | 0.7168472 |
| Normalized Enrichment Score (NES) | 2.050946 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0033908063 |
| FWER p-Value | 0.09 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | COL1A2 | COL1A2 Entrez, Source | collagen, type I, alpha 2 | 3 | 1.046 | 0.2667 | Yes |
| 2 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 32 | 0.660 | 0.4330 | Yes |
| 3 | TIMP2 | TIMP2 Entrez, Source | TIMP metallopeptidase inhibitor 2 | 35 | 0.642 | 0.5966 | Yes |
| 4 | CD9 | CD9 Entrez, Source | CD9 molecule | 299 | 0.250 | 0.6408 | Yes |
| 5 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 496 | 0.183 | 0.6727 | Yes |
| 6 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 717 | 0.149 | 0.6942 | Yes |
| 7 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 866 | 0.132 | 0.7168 | Yes |
| 8 | BAD | BAD Entrez, Source | BCL2-antagonist of cell death | 1750 | 0.083 | 0.6716 | No |
| 9 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 2079 | 0.071 | 0.6650 | No |
| 10 | CAV2 | CAV2 Entrez, Source | caveolin 2 | 2151 | 0.069 | 0.6771 | No |
| 11 | ANK1 | ANK1 Entrez, Source | ankyrin 1, erythrocytic | 2657 | 0.056 | 0.6535 | No |
| 12 | RHOG | RHOG Entrez, Source | ras homolog gene family, member G (rho G) | 2819 | 0.052 | 0.6547 | No |
| 13 | NGFR | NGFR Entrez, Source | nerve growth factor receptor (TNFR superfamily, member 16) | 2868 | 0.051 | 0.6643 | No |
| 14 | PDGFRB | PDGFRB Entrez, Source | platelet-derived growth factor receptor, beta polypeptide | 2871 | 0.051 | 0.6772 | No |
| 15 | BSG | BSG Entrez, Source | basigin (Ok blood group) | 3928 | 0.031 | 0.6059 | No |
| 16 | LUM | LUM Entrez, Source | lumican | 4728 | 0.019 | 0.5508 | No |
| 17 | ARHGDIB | ARHGDIB Entrez, Source | Rho GDP dissociation inhibitor (GDI) beta | 5075 | 0.014 | 0.5285 | No |
| 18 | VTN | VTN Entrez, Source | vitronectin | 5129 | 0.014 | 0.5280 | No |
| 19 | QSCN6 | QSCN6 Entrez, Source | quiescin Q6 | 5385 | 0.010 | 0.5114 | No |
| 20 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 6239 | -0.001 | 0.4476 | No |
| 21 | CASP9 | CASP9 Entrez, Source | caspase 9, apoptosis-related cysteine peptidase | 6766 | -0.008 | 0.4101 | No |
| 22 | MMP11 | MMP11 Entrez, Source | matrix metallopeptidase 11 (stromelysin 3) | 6969 | -0.010 | 0.3976 | No |
| 23 | FASTK | FASTK Entrez, Source | Fas-activated serine/threonine kinase | 7022 | -0.011 | 0.3964 | No |
| 24 | CD8B | CD8B Entrez, Source | CD8b molecule | 8424 | -0.030 | 0.2988 | No |
| 25 | PLA2G6 | PLA2G6 Entrez, Source | phospholipase A2, group VI (cytosolic, calcium-independent) | 8737 | -0.034 | 0.2841 | No |
| 26 | ITGA7 | ITGA7 Entrez, Source | integrin, alpha 7 | 9373 | -0.043 | 0.2473 | No |
| 27 | JUP | JUP Entrez, Source | junction plakoglobin | 10241 | -0.057 | 0.1968 | No |
| 28 | FBN2 | FBN2 Entrez, Source | fibrillin 2 (congenital contractural arachnodactyly) | 10607 | -0.064 | 0.1859 | No |
| 29 | IGFBP2 | IGFBP2 Entrez, Source | insulin-like growth factor binding protein 2, 36kDa | 11121 | -0.076 | 0.1668 | No |