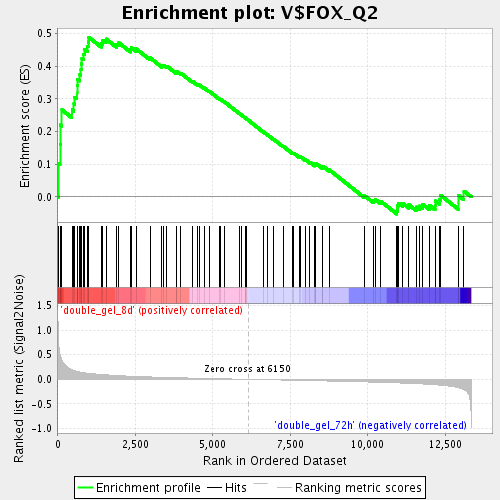
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | V$FOX_Q2 |
| Enrichment Score (ES) | 0.48881665 |
| Normalized Enrichment Score (NES) | 1.7478632 |
| Nominal p-value | 0.0016750419 |
| FDR q-value | 0.04164429 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 11 | 0.815 | 0.1030 | Yes |
| 2 | PRKAR2B | PRKAR2B Entrez, Source | protein kinase, cAMP-dependent, regulatory, type II, beta | 69 | 0.494 | 0.1615 | Yes |
| 3 | FSTL1 | FSTL1 Entrez, Source | follistatin-like 1 | 80 | 0.469 | 0.2205 | Yes |
| 4 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 130 | 0.390 | 0.2665 | Yes |
| 5 | MGLL | MGLL Entrez, Source | monoglyceride lipase | 454 | 0.195 | 0.2669 | Yes |
| 6 | IGSF4 | IGSF4 Entrez, Source | immunoglobulin superfamily, member 4 | 517 | 0.180 | 0.2851 | Yes |
| 7 | SLC20A1 | SLC20A1 Entrez, Source | solute carrier family 20 (phosphate transporter), member 1 | 543 | 0.173 | 0.3052 | Yes |
| 8 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 616 | 0.163 | 0.3205 | Yes |
| 9 | TGIF | TGIF Entrez, Source | TGFB-induced factor (TALE family homeobox) | 618 | 0.162 | 0.3412 | Yes |
| 10 | FABP4 | FABP4 Entrez, Source | fatty acid binding protein 4, adipocyte | 638 | 0.159 | 0.3600 | Yes |
| 11 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 708 | 0.150 | 0.3738 | Yes |
| 12 | NFIA | NFIA Entrez, Source | nuclear factor I/A | 743 | 0.145 | 0.3897 | Yes |
| 13 | PLAG1 | PLAG1 Entrez, Source | pleiomorphic adenoma gene 1 | 756 | 0.144 | 0.4072 | Yes |
| 14 | NCDN | NCDN Entrez, Source | neurochondrin | 774 | 0.142 | 0.4240 | Yes |
| 15 | LMO4 | LMO4 Entrez, Source | LIM domain only 4 | 827 | 0.136 | 0.4374 | Yes |
| 16 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 866 | 0.132 | 0.4514 | Yes |
| 17 | PALMD | PALMD Entrez, Source | palmdelphin | 958 | 0.125 | 0.4604 | Yes |
| 18 | TWIST1 | TWIST1 Entrez, Source | twist homolog 1 (acrocephalosyndactyly 3; Saethre-Chotzen syndrome) (Drosophila) | 978 | 0.123 | 0.4747 | Yes |
| 19 | GFRA1 | GFRA1 Entrez, Source | GDNF family receptor alpha 1 | 997 | 0.122 | 0.4888 | Yes |
| 20 | ING3 | ING3 Entrez, Source | inhibitor of growth family, member 3 | 1398 | 0.099 | 0.4712 | No |
| 21 | ATP1B4 | ATP1B4 Entrez, Source | ATPase, (Na+)/K+ transporting, beta 4 polypeptide | 1447 | 0.096 | 0.4798 | No |
| 22 | EIF4EBP2 | EIF4EBP2 Entrez, Source | eukaryotic translation initiation factor 4E binding protein 2 | 1553 | 0.091 | 0.4835 | No |
| 23 | SCG3 | SCG3 Entrez, Source | secretogranin III | 1896 | 0.077 | 0.4675 | No |
| 24 | NRG1 | NRG1 Entrez, Source | neuregulin 1 | 1967 | 0.074 | 0.4717 | No |
| 25 | CTHRC1 | CTHRC1 Entrez, Source | collagen triple helix repeat containing 1 | 2341 | 0.063 | 0.4516 | No |
| 26 | LGI1 | LGI1 Entrez, Source | leucine-rich, glioma inactivated 1 | 2365 | 0.063 | 0.4579 | No |
| 27 | TTR | TTR Entrez, Source | transthyretin (prealbumin, amyloidosis type I) | 2525 | 0.059 | 0.4534 | No |
| 28 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 2974 | 0.049 | 0.4259 | No |
| 29 | CDH10 | CDH10 Entrez, Source | cadherin 10, type 2 (T2-cadherin) | 3356 | 0.041 | 0.4024 | No |
| 30 | GDNF | GDNF Entrez, Source | glial cell derived neurotrophic factor | 3413 | 0.040 | 0.4033 | No |
| 31 | PHEX | PHEX Entrez, Source | phosphate regulating endopeptidase homolog, X-linked (hypophosphatemia, vitamin D resistant rickets) | 3518 | 0.038 | 0.4003 | No |
| 32 | TWIST2 | TWIST2 Entrez, Source | twist homolog 2 (Drosophila) | 3829 | 0.033 | 0.3811 | No |
| 33 | NEO1 | NEO1 Entrez, Source | neogenin homolog 1 (chicken) | 3835 | 0.033 | 0.3849 | No |
| 34 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 3967 | 0.031 | 0.3790 | No |
| 35 | TPP2 | TPP2 Entrez, Source | tripeptidyl peptidase II | 4336 | 0.025 | 0.3544 | No |
| 36 | GNAO1 | GNAO1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O | 4514 | 0.022 | 0.3439 | No |
| 37 | GPR1 | GPR1 Entrez, Source | G protein-coupled receptor 1 | 4565 | 0.022 | 0.3428 | No |
| 38 | VAX1 | VAX1 Entrez, Source | ventral anterior homeobox 1 | 4721 | 0.019 | 0.3336 | No |
| 39 | CYP26B1 | CYP26B1 Entrez, Source | cytochrome P450, family 26, subfamily B, polypeptide 1 | 4879 | 0.017 | 0.3239 | No |
| 40 | HTR1B | HTR1B Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 1B | 5221 | 0.012 | 0.2998 | No |
| 41 | NCAM1 | NCAM1 Entrez, Source | neural cell adhesion molecule 1 | 5249 | 0.012 | 0.2992 | No |
| 42 | ESRRG | ESRRG Entrez, Source | estrogen-related receptor gamma | 5378 | 0.010 | 0.2909 | No |
| 43 | WBSCR1 | WBSCR1 Entrez, Source | Williams-Beuren syndrome chromosome region 1 | 5863 | 0.004 | 0.2548 | No |
| 44 | GPRC5C | GPRC5C Entrez, Source | G protein-coupled receptor, family C, group 5, member C | 5932 | 0.003 | 0.2501 | No |
| 45 | ICK | ICK Entrez, Source | intestinal cell (MAK-like) kinase | 6039 | 0.002 | 0.2423 | No |
| 46 | NTN1 | NTN1 Entrez, Source | netrin 1 | 6053 | 0.001 | 0.2415 | No |
| 47 | PVRL1 | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 6070 | 0.001 | 0.2405 | No |
| 48 | PPP1CB | PPP1CB Entrez, Source | protein phosphatase 1, catalytic subunit, beta isoform | 6632 | -0.006 | 0.1989 | No |
| 49 | NEUROD2 | NEUROD2 Entrez, Source | neurogenic differentiation 2 | 6765 | -0.008 | 0.1900 | No |
| 50 | SMARCA2 | SMARCA2 Entrez, Source | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 6971 | -0.010 | 0.1758 | No |
| 51 | ARRB2 | ARRB2 Entrez, Source | arrestin, beta 2 | 7277 | -0.014 | 0.1546 | No |
| 52 | SLIT3 | SLIT3 Entrez, Source | slit homolog 3 (Drosophila) | 7586 | -0.018 | 0.1336 | No |
| 53 | RFX3 | RFX3 Entrez, Source | regulatory factor X, 3 (influences HLA class II expression) | 7612 | -0.018 | 0.1341 | No |
| 54 | NTF3 | NTF3 Entrez, Source | neurotrophin 3 | 7788 | -0.021 | 0.1235 | No |
| 55 | FKBP14 | FKBP14 Entrez, Source | FK506 binding protein 14, 22 kDa | 7815 | -0.021 | 0.1242 | No |
| 56 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 7976 | -0.023 | 0.1151 | No |
| 57 | SLC39A8 | SLC39A8 Entrez, Source | solute carrier family 39 (zinc transporter), member 8 | 8133 | -0.026 | 0.1066 | No |
| 58 | PRDX5 | PRDX5 Entrez, Source | peroxiredoxin 5 | 8294 | -0.028 | 0.0980 | No |
| 59 | H2AFY | H2AFY Entrez, Source | H2A histone family, member Y | 8311 | -0.028 | 0.1004 | No |
| 60 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 8322 | -0.028 | 0.1032 | No |
| 61 | BAMBI | BAMBI Entrez, Source | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | 8540 | -0.031 | 0.0908 | No |
| 62 | MAP2K5 | MAP2K5 Entrez, Source | mitogen-activated protein kinase kinase 5 | 8547 | -0.031 | 0.0944 | No |
| 63 | CASQ2 | CASQ2 Entrez, Source | calsequestrin 2 (cardiac muscle) | 8759 | -0.034 | 0.0828 | No |
| 64 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 9899 | -0.052 | 0.0035 | No |
| 65 | ARHGEF2 | ARHGEF2 Entrez, Source | rho/rac guanine nucleotide exchange factor (GEF) 2 | 10190 | -0.057 | -0.0112 | No |
| 66 | GRIK3 | GRIK3 Entrez, Source | glutamate receptor, ionotropic, kainate 3 | 10240 | -0.057 | -0.0075 | No |
| 67 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10414 | -0.060 | -0.0129 | No |
| 68 | STOML2 | STOML2 Entrez, Source | stomatin (EPB72)-like 2 | 10942 | -0.072 | -0.0435 | No |
| 69 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 10957 | -0.072 | -0.0353 | No |
| 70 | PDCD4 | PDCD4 Entrez, Source | programmed cell death 4 (neoplastic transformation inhibitor) | 10963 | -0.073 | -0.0265 | No |
| 71 | RAB3IP | RAB3IP Entrez, Source | RAB3A interacting protein (rabin3) | 10986 | -0.073 | -0.0188 | No |
| 72 | TNR | TNR Entrez, Source | tenascin R (restrictin, janusin) | 11126 | -0.076 | -0.0196 | No |
| 73 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 11328 | -0.082 | -0.0243 | No |
| 74 | RPS6KB1 | RPS6KB1 Entrez, Source | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | 11564 | -0.088 | -0.0308 | No |
| 75 | KCNH5 | KCNH5 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 5 | 11678 | -0.091 | -0.0277 | No |
| 76 | FBXO11 | FBXO11 Entrez, Source | F-box protein 11 | 11774 | -0.094 | -0.0229 | No |
| 77 | EFNA1 | EFNA1 Entrez, Source | ephrin-A1 | 11985 | -0.102 | -0.0257 | No |
| 78 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 12174 | -0.110 | -0.0258 | No |
| 79 | EFEMP1 | EFEMP1 Entrez, Source | EGF-containing fibulin-like extracellular matrix protein 1 | 12182 | -0.111 | -0.0123 | No |
| 80 | FGF13 | FGF13 Entrez, Source | fibroblast growth factor 13 | 12314 | -0.117 | -0.0073 | No |
| 81 | GATA6 | GATA6 Entrez, Source | GATA binding protein 6 | 12352 | -0.119 | 0.0050 | No |
| 82 | CADPS | CADPS Entrez, Source | Ca2+-dependent secretion activator | 12921 | -0.167 | -0.0165 | No |
| 83 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 12922 | -0.168 | 0.0048 | No |
| 84 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 13105 | -0.210 | 0.0179 | No |