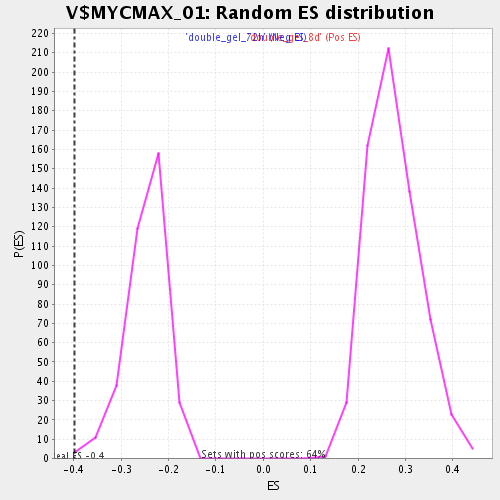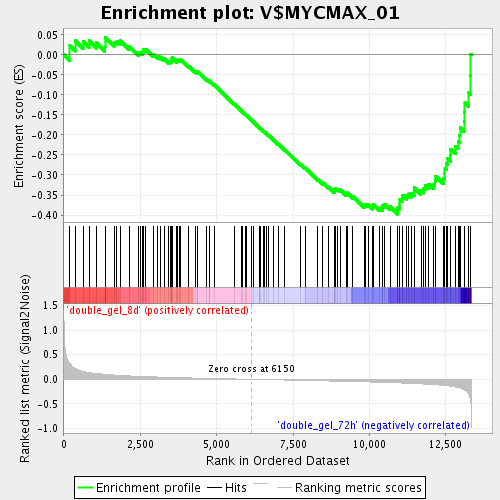
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | V$MYCMAX_01 |
| Enrichment Score (ES) | -0.39761543 |
| Normalized Enrichment Score (NES) | -1.6059135 |
| Nominal p-value | 0.005586592 |
| FDR q-value | 0.2129015 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 196 | 0.324 | 0.0224 | No |
| 2 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 367 | 0.223 | 0.0351 | No |
| 3 | KCNH4 | KCNH4 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 4 | 628 | 0.161 | 0.0340 | No |
| 4 | DIRC2 | DIRC2 Entrez, Source | disrupted in renal carcinoma 2 | 823 | 0.137 | 0.0351 | No |
| 5 | TRIB1 | TRIB1 Entrez, Source | tribbles homolog 1 (Drosophila) | 1067 | 0.116 | 0.0301 | No |
| 6 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 1344 | 0.102 | 0.0209 | No |
| 7 | PER1 | PER1 Entrez, Source | period homolog 1 (Drosophila) | 1347 | 0.101 | 0.0324 | No |
| 8 | CRMP1 | CRMP1 Entrez, Source | collapsin response mediator protein 1 | 1349 | 0.101 | 0.0439 | No |
| 9 | RAMP2 | RAMP2 Entrez, Source | receptor (calcitonin) activity modifying protein 2 | 1641 | 0.087 | 0.0320 | No |
| 10 | G6PC3 | G6PC3 Entrez, Source | glucose 6 phosphatase, catalytic, 3 | 1736 | 0.083 | 0.0344 | No |
| 11 | BMP4 | BMP4 Entrez, Source | bone morphogenetic protein 4 | 1840 | 0.078 | 0.0356 | No |
| 12 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 2135 | 0.069 | 0.0213 | No |
| 13 | DIABLO | DIABLO Entrez, Source | diablo homolog (Drosophila) | 2448 | 0.061 | 0.0047 | No |
| 14 | GPC3 | GPC3 Entrez, Source | glypican 3 | 2514 | 0.059 | 0.0066 | No |
| 15 | SLC6A15 | SLC6A15 Entrez, Source | solute carrier family 6, member 15 | 2585 | 0.058 | 0.0080 | No |
| 16 | RHEBL1 | RHEBL1 Entrez, Source | Ras homolog enriched in brain like 1 | 2608 | 0.057 | 0.0129 | No |
| 17 | RAB3IL1 | RAB3IL1 Entrez, Source | RAB3A interacting protein (rabin3)-like 1 | 2673 | 0.056 | 0.0145 | No |
| 18 | GIT1 | GIT1 Entrez, Source | G protein-coupled receptor kinase interactor 1 | 2932 | 0.050 | 0.0007 | No |
| 19 | MPP3 | MPP3 Entrez, Source | membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | 3078 | 0.047 | -0.0048 | No |
| 20 | SUCLG2 | SUCLG2 Entrez, Source | succinate-CoA ligase, GDP-forming, beta subunit | 3155 | 0.045 | -0.0054 | No |
| 21 | BOK | BOK Entrez, Source | BCL2-related ovarian killer | 3278 | 0.043 | -0.0096 | No |
| 22 | TUBA4 | TUBA4 Entrez, Source | tubulin, alpha 4 | 3435 | 0.040 | -0.0169 | No |
| 23 | ELAVL3 | ELAVL3 Entrez, Source | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) | 3483 | 0.039 | -0.0159 | No |
| 24 | POU3F2 | POU3F2 Entrez, Source | POU domain, class 3, transcription factor 2 | 3514 | 0.038 | -0.0138 | No |
| 25 | CSK | CSK Entrez, Source | c-src tyrosine kinase | 3515 | 0.038 | -0.0094 | No |
| 26 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 3538 | 0.038 | -0.0067 | No |
| 27 | LZTS2 | LZTS2 Entrez, Source | leucine zipper, putative tumor suppressor 2 | 3697 | 0.035 | -0.0146 | No |
| 28 | PPP1R9B | PPP1R9B Entrez, Source | protein phosphatase 1, regulatory subunit 9B, spinophilin | 3726 | 0.035 | -0.0127 | No |
| 29 | EFNB1 | EFNB1 Entrez, Source | ephrin-B1 | 3795 | 0.034 | -0.0140 | No |
| 30 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 3800 | 0.033 | -0.0105 | No |
| 31 | SC65 | SC65 Entrez, Source | - | 4076 | 0.029 | -0.0279 | No |
| 32 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 4293 | 0.026 | -0.0413 | No |
| 33 | STXBP2 | STXBP2 Entrez, Source | syntaxin binding protein 2 | 4370 | 0.024 | -0.0443 | No |
| 34 | PPARGC1B | PPARGC1B Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, beta | 4372 | 0.024 | -0.0416 | No |
| 35 | U2AF2 | U2AF2 Entrez, Source | U2 small nuclear RNA auxiliary factor 2 | 4679 | 0.020 | -0.0624 | No |
| 36 | ARL3 | ARL3 Entrez, Source | ADP-ribosylation factor-like 3 | 4754 | 0.019 | -0.0658 | No |
| 37 | CGREF1 | CGREF1 Entrez, Source | cell growth regulator with EF-hand domain 1 | 4769 | 0.019 | -0.0647 | No |
| 38 | FOXD3 | FOXD3 Entrez, Source | forkhead box D3 | 4916 | 0.016 | -0.0739 | No |
| 39 | COL2A1 | COL2A1 Entrez, Source | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 5569 | 0.007 | -0.1223 | No |
| 40 | GPS1 | GPS1 Entrez, Source | G protein pathway suppressor 1 | 5817 | 0.004 | -0.1405 | No |
| 41 | HNRPDL | HNRPDL Entrez, Source | heterogeneous nuclear ribonucleoprotein D-like | 5830 | 0.004 | -0.1410 | No |
| 42 | GPRC5C | GPRC5C Entrez, Source | G protein-coupled receptor, family C, group 5, member C | 5932 | 0.003 | -0.1483 | No |
| 43 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 5974 | 0.002 | -0.1511 | No |
| 44 | SYT3 | SYT3 Entrez, Source | synaptotagmin III | 6132 | 0.000 | -0.1629 | No |
| 45 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 6189 | -0.000 | -0.1671 | No |
| 46 | PITX3 | PITX3 Entrez, Source | paired-like homeodomain transcription factor 3 | 6413 | -0.003 | -0.1836 | No |
| 47 | ESRRA | ESRRA Entrez, Source | estrogen-related receptor alpha | 6415 | -0.003 | -0.1833 | No |
| 48 | KCMF1 | KCMF1 Entrez, Source | potassium channel modulatory factor 1 | 6430 | -0.004 | -0.1839 | No |
| 49 | NRAS | NRAS Entrez, Source | neuroblastoma RAS viral (v-ras) oncogene homolog | 6534 | -0.005 | -0.1911 | No |
| 50 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 6574 | -0.005 | -0.1935 | No |
| 51 | RUNX2 | RUNX2 Entrez, Source | runt-related transcription factor 2 | 6625 | -0.006 | -0.1966 | No |
| 52 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 6700 | -0.007 | -0.2013 | No |
| 53 | CLN3 | CLN3 Entrez, Source | ceroid-lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 6869 | -0.009 | -0.2130 | No |
| 54 | FGF14 | FGF14 Entrez, Source | fibroblast growth factor 14 | 7024 | -0.011 | -0.2234 | No |
| 55 | PFN1 | PFN1 Entrez, Source | profilin 1 | 7229 | -0.013 | -0.2372 | No |
| 56 | QTRT1 | QTRT1 Entrez, Source | queuine tRNA-ribosyltransferase 1 (tRNA-guanine transglycosylase) | 7751 | -0.020 | -0.2743 | No |
| 57 | FGF6 | FGF6 Entrez, Source | fibroblast growth factor 6 | 7892 | -0.022 | -0.2824 | No |
| 58 | LYAR | LYAR Entrez, Source | - | 8301 | -0.028 | -0.3100 | No |
| 59 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 8470 | -0.031 | -0.3192 | No |
| 60 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 8663 | -0.033 | -0.3299 | No |
| 61 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 8845 | -0.035 | -0.3395 | No |
| 62 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 8847 | -0.035 | -0.3356 | No |
| 63 | ACY1 | ACY1 Entrez, Source | aminoacylase 1 | 8889 | -0.036 | -0.3345 | No |
| 64 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 8961 | -0.037 | -0.3356 | No |
| 65 | NUDC | NUDC Entrez, Source | nuclear distribution gene C homolog (A. nidulans) | 9037 | -0.038 | -0.3369 | No |
| 66 | OPRS1 | OPRS1 Entrez, Source | opioid receptor, sigma 1 | 9241 | -0.041 | -0.3476 | No |
| 67 | ENO3 | ENO3 Entrez, Source | enolase 3 (beta, muscle) | 9266 | -0.041 | -0.3447 | No |
| 68 | NCL | NCL Entrez, Source | nucleolin | 9448 | -0.044 | -0.3533 | No |
| 69 | PSME3 | PSME3 Entrez, Source | proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) | 9831 | -0.051 | -0.3763 | No |
| 70 | WDR23 | WDR23 Entrez, Source | WD repeat domain 23 | 9861 | -0.051 | -0.3726 | No |
| 71 | MRPL40 | MRPL40 Entrez, Source | mitochondrial ribosomal protein L40 | 9957 | -0.053 | -0.3738 | No |
| 72 | MEOX2 | MEOX2 Entrez, Source | mesenchyme homeobox 2 | 10109 | -0.055 | -0.3788 | No |
| 73 | SLC12A5 | SLC12A5 Entrez, Source | solute carrier family 12, (potassium-chloride transporter) member 5 | 10117 | -0.055 | -0.3730 | No |
| 74 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 10331 | -0.059 | -0.3823 | No |
| 75 | EWSR1 | EWSR1 Entrez, Source | Ewing sarcoma breakpoint region 1 | 10419 | -0.061 | -0.3819 | No |
| 76 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 10434 | -0.061 | -0.3760 | No |
| 77 | OPRD1 | OPRD1 Entrez, Source | opioid receptor, delta 1 | 10491 | -0.062 | -0.3731 | No |
| 78 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 10672 | -0.066 | -0.3792 | No |
| 79 | RAI14 | RAI14 Entrez, Source | retinoic acid induced 14 | 10917 | -0.071 | -0.3894 | Yes |
| 80 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 10929 | -0.072 | -0.3820 | Yes |
| 81 | PABPC1 | PABPC1 Entrez, Source | poly(A) binding protein, cytoplasmic 1 | 10990 | -0.073 | -0.3782 | Yes |
| 82 | RCL1 | RCL1 Entrez, Source | RNA terminal phosphate cyclase-like 1 | 10995 | -0.073 | -0.3701 | Yes |
| 83 | EIF2C2 | EIF2C2 Entrez, Source | eukaryotic translation initiation factor 2C, 2 | 10997 | -0.073 | -0.3617 | Yes |
| 84 | PUS1 | PUS1 Entrez, Source | pseudouridylate synthase 1 | 11069 | -0.075 | -0.3585 | Yes |
| 85 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 11090 | -0.075 | -0.3513 | Yes |
| 86 | IPO13 | IPO13 Entrez, Source | importin 13 | 11209 | -0.079 | -0.3512 | Yes |
| 87 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 11282 | -0.081 | -0.3474 | Yes |
| 88 | TESK2 | TESK2 Entrez, Source | testis-specific kinase 2 | 11384 | -0.084 | -0.3454 | Yes |
| 89 | KCNN4 | KCNN4 Entrez, Source | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 11462 | -0.085 | -0.3414 | Yes |
| 90 | GPM6B | GPM6B Entrez, Source | glycoprotein M6B | 11468 | -0.085 | -0.3320 | Yes |
| 91 | USP15 | USP15 Entrez, Source | ubiquitin specific peptidase 15 | 11686 | -0.091 | -0.3379 | Yes |
| 92 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 11781 | -0.095 | -0.3341 | Yes |
| 93 | NOL5A | NOL5A Entrez, Source | nucleolar protein 5A (56kDa with KKE/D repeat) | 11821 | -0.096 | -0.3260 | Yes |
| 94 | TIMM8A | TIMM8A Entrez, Source | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 11939 | -0.100 | -0.3233 | Yes |
| 95 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 12093 | -0.107 | -0.3226 | Yes |
| 96 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 12144 | -0.109 | -0.3139 | Yes |
| 97 | HTF9C | HTF9C Entrez, Source | - | 12171 | -0.110 | -0.3032 | Yes |
| 98 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 12425 | -0.123 | -0.3082 | Yes |
| 99 | HSPBAP1 | HSPBAP1 Entrez, Source | HSPB (heat shock 27kDa) associated protein 1 | 12466 | -0.125 | -0.2969 | Yes |
| 100 | RPS19 | RPS19 Entrez, Source | ribosomal protein S19 | 12470 | -0.125 | -0.2827 | Yes |
| 101 | ATAD3A | ATAD3A Entrez, Source | ATPase family, AAA domain containing 3A | 12521 | -0.130 | -0.2716 | Yes |
| 102 | NEUROD1 | NEUROD1 Entrez, Source | neurogenic differentiation 1 | 12563 | -0.132 | -0.2595 | Yes |
| 103 | ILF3 | ILF3 Entrez, Source | interleukin enhancer binding factor 3, 90kDa | 12652 | -0.138 | -0.2503 | Yes |
| 104 | FKBP11 | FKBP11 Entrez, Source | FK506 binding protein 11, 19 kDa | 12662 | -0.138 | -0.2351 | Yes |
| 105 | PPAT | PPAT Entrez, Source | phosphoribosyl pyrophosphate amidotransferase | 12816 | -0.154 | -0.2289 | Yes |
| 106 | XTP3TPA | XTP3TPA Entrez, Source | - | 12898 | -0.165 | -0.2162 | Yes |
| 107 | ZBTB10 | ZBTB10 Entrez, Source | zinc finger and BTB domain containing 10 | 12951 | -0.173 | -0.2002 | Yes |
| 108 | LAP3 | LAP3 Entrez, Source | leucine aminopeptidase 3 | 12964 | -0.175 | -0.1810 | Yes |
| 109 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 13096 | -0.208 | -0.1670 | Yes |
| 110 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 13105 | -0.210 | -0.1434 | Yes |
| 111 | DAZAP1 | DAZAP1 Entrez, Source | DAZ associated protein 1 | 13125 | -0.217 | -0.1200 | Yes |
| 112 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 13254 | -0.302 | -0.0949 | Yes |
| 113 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 13302 | -0.408 | -0.0516 | Yes |
| 114 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 13317 | -0.475 | 0.0019 | Yes |