Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | VENTRICLES_UP |
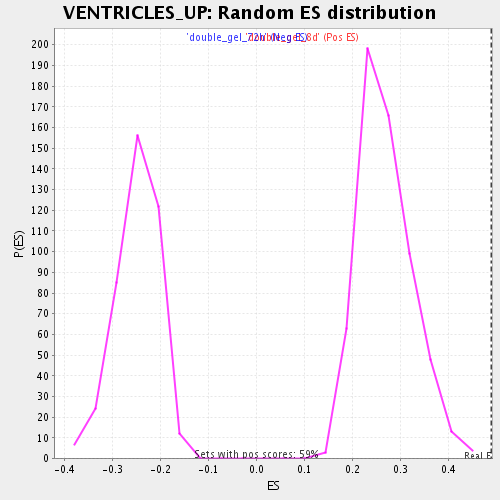
| Enrichment Score (ES) | 0.4898289 |
| Normalized Enrichment Score (NES) | 1.8218982 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.029448539 |
| FWER p-Value | 0.938 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 11 | 0.815 | 0.0710 | Yes |
| 2 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 22 | 0.685 | 0.1306 | Yes |
| 3 | LPL | LPL Entrez, Source | lipoprotein lipase | 24 | 0.680 | 0.1904 | Yes |
| 4 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 52 | 0.576 | 0.2391 | Yes |
| 5 | CDH13 | CDH13 Entrez, Source | cadherin 13, H-cadherin (heart) | 128 | 0.391 | 0.2678 | Yes |
| 6 | PLCL1 | PLCL1 Entrez, Source | phospholipase C-like 1 | 169 | 0.348 | 0.2955 | Yes |
| 7 | SLC22A5 | SLC22A5 Entrez, Source | solute carrier family 22 (organic cation transporter), member 5 | 204 | 0.319 | 0.3210 | Yes |
| 8 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 222 | 0.305 | 0.3466 | Yes |
| 9 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 528 | 0.177 | 0.3391 | Yes |
| 10 | SLC25A20 | SLC25A20 Entrez, Source | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 | 636 | 0.159 | 0.3450 | Yes |
| 11 | ITGA9 | ITGA9 Entrez, Source | integrin, alpha 9 | 643 | 0.158 | 0.3585 | Yes |
| 12 | FABP3 | FABP3 Entrez, Source | fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 666 | 0.155 | 0.3705 | Yes |
| 13 | NDRG4 | NDRG4 Entrez, Source | NDRG family member 4 | 677 | 0.153 | 0.3832 | Yes |
| 14 | LMO7 | LMO7 Entrez, Source | LIM domain 7 | 728 | 0.147 | 0.3924 | Yes |
| 15 | IMPA2 | IMPA2 Entrez, Source | inositol(myo)-1(or 4)-monophosphatase 2 | 735 | 0.146 | 0.4048 | Yes |
| 16 | STK39 | STK39 Entrez, Source | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | 864 | 0.133 | 0.4068 | Yes |
| 17 | GPC1 | GPC1 Entrez, Source | glypican 1 | 929 | 0.127 | 0.4132 | Yes |
| 18 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 930 | 0.126 | 0.4243 | Yes |
| 19 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 1098 | 0.115 | 0.4218 | Yes |
| 20 | HSPB6 | HSPB6 Entrez, Source | heat shock protein, alpha-crystallin-related, B6 | 1106 | 0.114 | 0.4314 | Yes |
| 21 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 1177 | 0.111 | 0.4358 | Yes |
| 22 | SORBS2 | SORBS2 Entrez, Source | sorbin and SH3 domain containing 2 | 1187 | 0.110 | 0.4448 | Yes |
| 23 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 1245 | 0.108 | 0.4500 | Yes |
| 24 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1289 | 0.105 | 0.4560 | Yes |
| 25 | KCNQ1 | KCNQ1 Entrez, Source | potassium voltage-gated channel, KQT-like subfamily, member 1 | 1386 | 0.099 | 0.4574 | Yes |
| 26 | UBE1C | UBE1C Entrez, Source | ubiquitin-activating enzyme E1C (UBA3 homolog, yeast) | 1486 | 0.095 | 0.4583 | Yes |
| 27 | PDLIM1 | PDLIM1 Entrez, Source | PDZ and LIM domain 1 (elfin) | 1487 | 0.095 | 0.4666 | Yes |
| 28 | CHN1 | CHN1 Entrez, Source | chimerin (chimaerin) 1 | 1627 | 0.088 | 0.4639 | Yes |
| 29 | APOD | APOD Entrez, Source | apolipoprotein D | 1653 | 0.087 | 0.4696 | Yes |
| 30 | KIFC3 | KIFC3 Entrez, Source | kinesin family member C3 | 1724 | 0.084 | 0.4717 | Yes |
| 31 | CAPN2 | CAPN2 Entrez, Source | calpain 2, (m/II) large subunit | 1726 | 0.084 | 0.4790 | Yes |
| 32 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 1791 | 0.081 | 0.4812 | Yes |
| 33 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1828 | 0.079 | 0.4855 | Yes |
| 34 | P2RY2 | P2RY2 Entrez, Source | purinergic receptor P2Y, G-protein coupled, 2 | 1862 | 0.078 | 0.4898 | Yes |
| 35 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 2119 | 0.069 | 0.4766 | No |
| 36 | CXADR | CXADR Entrez, Source | coxsackie virus and adenovirus receptor | 2231 | 0.066 | 0.4740 | No |
| 37 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 2265 | 0.065 | 0.4773 | No |
| 38 | NLGN1 | NLGN1 Entrez, Source | neuroligin 1 | 2284 | 0.065 | 0.4816 | No |
| 39 | KCNJ8 | KCNJ8 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 8 | 2439 | 0.061 | 0.4754 | No |
| 40 | ASS1 | ASS1 Entrez, Source | argininosuccinate synthetase 1 | 2650 | 0.056 | 0.4644 | No |
| 41 | PKIA | PKIA Entrez, Source | protein kinase (cAMP-dependent, catalytic) inhibitor alpha | 2727 | 0.054 | 0.4635 | No |
| 42 | APOA1 | APOA1 Entrez, Source | apolipoprotein A-I | 2953 | 0.050 | 0.4508 | No |
| 43 | SH3GL2 | SH3GL2 Entrez, Source | SH3-domain GRB2-like 2 | 2976 | 0.049 | 0.4535 | No |
| 44 | BCAT2 | BCAT2 Entrez, Source | branched chain aminotransferase 2, mitochondrial | 3239 | 0.044 | 0.4376 | No |
| 45 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 3326 | 0.042 | 0.4347 | No |
| 46 | GNAS | GNAS Entrez, Source | GNAS complex locus | 3379 | 0.041 | 0.4344 | No |
| 47 | GRM1 | GRM1 Entrez, Source | glutamate receptor, metabotropic 1 | 3497 | 0.039 | 0.4290 | No |
| 48 | PDE1C | PDE1C Entrez, Source | phosphodiesterase 1C, calmodulin-dependent 70kDa | 3643 | 0.036 | 0.4212 | No |
| 49 | PCSK6 | PCSK6 Entrez, Source | proprotein convertase subtilisin/kexin type 6 | 3695 | 0.035 | 0.4204 | No |
| 50 | COX8A | COX8A Entrez, Source | cytochrome c oxidase subunit 8A (ubiquitous) | 4033 | 0.030 | 0.3976 | No |
| 51 | PTPN3 | PTPN3 Entrez, Source | protein tyrosine phosphatase, non-receptor type 3 | 4204 | 0.027 | 0.3871 | No |
| 52 | CAPNS1 | CAPNS1 Entrez, Source | calpain, small subunit 1 | 4237 | 0.026 | 0.3870 | No |
| 53 | PAMCI | PAMCI Entrez, Source | peptidylglycine alpha-amidating monooxygenase COOH-terminal interactor | 4324 | 0.025 | 0.3827 | No |
| 54 | COX7A2 | COX7A2 Entrez, Source | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | 4338 | 0.025 | 0.3839 | No |
| 55 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 4734 | 0.019 | 0.3557 | No |
| 56 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 4788 | 0.018 | 0.3533 | No |
| 57 | DSCR1L1 | DSCR1L1 Entrez, Source | Down syndrome critical region gene 1-like 1 | 4800 | 0.018 | 0.3541 | No |
| 58 | ELAC2 | ELAC2 Entrez, Source | elaC homolog 2 (E. coli) | 4966 | 0.016 | 0.3430 | No |
| 59 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 4986 | 0.015 | 0.3429 | No |
| 60 | VTN | VTN Entrez, Source | vitronectin | 5129 | 0.014 | 0.3334 | No |
| 61 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 5319 | 0.011 | 0.3200 | No |
| 62 | MYL3 | MYL3 Entrez, Source | myosin, light chain 3, alkali; ventricular, skeletal, slow | 5408 | 0.010 | 0.3142 | No |
| 63 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 5585 | 0.007 | 0.3015 | No |
| 64 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 5788 | 0.005 | 0.2867 | No |
| 65 | CLCN1 | CLCN1 Entrez, Source | chloride channel 1, skeletal muscle (Thomsen disease, autosomal dominant) | 5834 | 0.004 | 0.2836 | No |
| 66 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 5958 | 0.003 | 0.2745 | No |
| 67 | CAP2 | CAP2 Entrez, Source | CAP, adenylate cyclase-associated protein, 2 (yeast) | 6076 | 0.001 | 0.2658 | No |
| 68 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 6135 | 0.000 | 0.2614 | No |
| 69 | SMYD2 | SMYD2 Entrez, Source | SET and MYND domain containing 2 | 6438 | -0.004 | 0.2389 | No |
| 70 | UBE2G1 | UBE2G1 Entrez, Source | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | 6486 | -0.004 | 0.2357 | No |
| 71 | RNF10 | RNF10 Entrez, Source | ring finger protein 10 | 6621 | -0.006 | 0.2261 | No |
| 72 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 6671 | -0.007 | 0.2230 | No |
| 73 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 6928 | -0.010 | 0.2045 | No |
| 74 | PKP2 | PKP2 Entrez, Source | plakophilin 2 | 7085 | -0.012 | 0.1937 | No |
| 75 | TJP2 | TJP2 Entrez, Source | tight junction protein 2 (zona occludens 2) | 7094 | -0.012 | 0.1941 | No |
| 76 | MAPKAPK3 | MAPKAPK3 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 3 | 7267 | -0.014 | 0.1823 | No |
| 77 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 7272 | -0.014 | 0.1833 | No |
| 78 | PHKA1 | PHKA1 Entrez, Source | phosphorylase kinase, alpha 1 (muscle) | 7383 | -0.015 | 0.1763 | No |
| 79 | PDE3A | PDE3A Entrez, Source | phosphodiesterase 3A, cGMP-inhibited | 7430 | -0.016 | 0.1742 | No |
| 80 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 7746 | -0.020 | 0.1521 | No |
| 81 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 7848 | -0.021 | 0.1464 | No |
| 82 | COPS3 | COPS3 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | 7930 | -0.022 | 0.1422 | No |
| 83 | NDUFS6 | NDUFS6 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) | 8128 | -0.025 | 0.1295 | No |
| 84 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 8238 | -0.027 | 0.1237 | No |
| 85 | COX5A | COX5A Entrez, Source | cytochrome c oxidase subunit Va | 8314 | -0.028 | 0.1205 | No |
| 86 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 8322 | -0.028 | 0.1224 | No |
| 87 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 8373 | -0.029 | 0.1212 | No |
| 88 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 8535 | -0.031 | 0.1117 | No |
| 89 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 8731 | -0.034 | 0.1000 | No |
| 90 | DLK1 | DLK1 Entrez, Source | delta-like 1 homolog (Drosophila) | 8739 | -0.034 | 0.1025 | No |
| 91 | OPA1 | OPA1 Entrez, Source | optic atrophy 1 (autosomal dominant) | 8921 | -0.036 | 0.0920 | No |
| 92 | NDUFV2 | NDUFV2 Entrez, Source | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | 9277 | -0.041 | 0.0688 | No |
| 93 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 9290 | -0.042 | 0.0715 | No |
| 94 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 9509 | -0.045 | 0.0590 | No |
| 95 | CDH2 | CDH2 Entrez, Source | cadherin 2, type 1, N-cadherin (neuronal) | 9642 | -0.048 | 0.0532 | No |
| 96 | PCYT2 | PCYT2 Entrez, Source | phosphate cytidylyltransferase 2, ethanolamine | 9674 | -0.048 | 0.0551 | No |
| 97 | FHL2 | FHL2 Entrez, Source | four and a half LIM domains 2 | 9790 | -0.050 | 0.0508 | No |
| 98 | NDUFA9 | NDUFA9 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 9915 | -0.052 | 0.0461 | No |
| 99 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 9920 | -0.052 | 0.0504 | No |
| 100 | UBADC1 | UBADC1 Entrez, Source | ubiquitin associated domain containing 1 | 10116 | -0.055 | 0.0405 | No |
| 101 | PGM1 | PGM1 Entrez, Source | phosphoglucomutase 1 | 10277 | -0.058 | 0.0335 | No |
| 102 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 10423 | -0.061 | 0.0279 | No |
| 103 | FGF18 | FGF18 Entrez, Source | fibroblast growth factor 18 | 10472 | -0.062 | 0.0297 | No |
| 104 | GBE1 | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 10501 | -0.062 | 0.0330 | No |
| 105 | PSMB6 | PSMB6 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 6 | 10550 | -0.063 | 0.0350 | No |
| 106 | PLN | PLN Entrez, Source | phospholamban | 10671 | -0.066 | 0.0317 | No |
| 107 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 10672 | -0.066 | 0.0375 | No |
| 108 | PDCD8 | PDCD8 Entrez, Source | programmed cell death 8 (apoptosis-inducing factor) | 10718 | -0.066 | 0.0399 | No |
| 109 | ALDH1L1 | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 10901 | -0.071 | 0.0324 | No |
| 110 | PTPRM | PTPRM Entrez, Source | protein tyrosine phosphatase, receptor type, M | 10978 | -0.073 | 0.0330 | No |
| 111 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 11025 | -0.074 | 0.0360 | No |
| 112 | CHN2 | CHN2 Entrez, Source | chimerin (chimaerin) 2 | 11079 | -0.075 | 0.0387 | No |
| 113 | RHOQ | RHOQ Entrez, Source | ras homolog gene family, member Q | 11086 | -0.075 | 0.0448 | No |
| 114 | DPYSL4 | DPYSL4 Entrez, Source | dihydropyrimidinase-like 4 | 11404 | -0.084 | 0.0283 | No |
| 115 | BDH1 | BDH1 Entrez, Source | 3-hydroxybutyrate dehydrogenase, type 1 | 11512 | -0.087 | 0.0278 | No |
| 116 | MYL2 | MYL2 Entrez, Source | myosin, light chain 2, regulatory, cardiac, slow | 11531 | -0.087 | 0.0341 | No |
| 117 | RAB21 | RAB21 Entrez, Source | RAB21, member RAS oncogene family | 11632 | -0.090 | 0.0345 | No |
| 118 | GABRE | GABRE Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, epsilon | 11946 | -0.101 | 0.0197 | No |
| 119 | PODXL | PODXL Entrez, Source | podocalyxin-like | 12132 | -0.109 | 0.0152 | No |
| 120 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 12195 | -0.111 | 0.0203 | No |
| 121 | AFG3L2 | AFG3L2 Entrez, Source | AFG3 ATPase family gene 3-like 2 (yeast) | 12218 | -0.112 | 0.0286 | No |
| 122 | HIPK3 | HIPK3 Entrez, Source | homeodomain interacting protein kinase 3 | 12735 | -0.145 | 0.0023 | No |
| 123 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 12987 | -0.180 | -0.0008 | No |
| 124 | CSRP2 | CSRP2 Entrez, Source | cysteine and glycine-rich protein 2 | 13262 | -0.313 | 0.0061 | No |