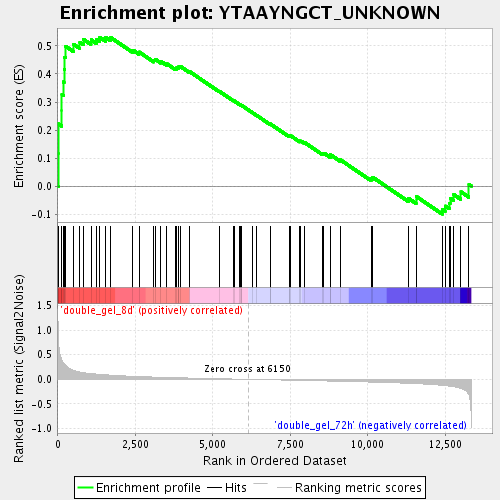
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | YTAAYNGCT_UNKNOWN |
| Enrichment Score (ES) | 0.53087103 |
| Normalized Enrichment Score (NES) | 1.7730968 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.036167607 |
| FWER p-Value | 0.994 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 11 | 0.815 | 0.1172 | Yes |
| 2 | DBP | DBP Entrez, Source | D site of albumin promoter (albumin D-box) binding protein | 16 | 0.727 | 0.2221 | Yes |
| 3 | CDH13 | CDH13 Entrez, Source | cadherin 13, H-cadherin (heart) | 128 | 0.391 | 0.2703 | Yes |
| 4 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 130 | 0.390 | 0.3267 | Yes |
| 5 | AMPD3 | AMPD3 Entrez, Source | adenosine monophosphate deaminase (isoform E) | 180 | 0.337 | 0.3718 | Yes |
| 6 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 201 | 0.321 | 0.4167 | Yes |
| 7 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 218 | 0.309 | 0.4602 | Yes |
| 8 | PEA15 | PEA15 Entrez, Source | phosphoprotein enriched in astrocytes 15 | 252 | 0.281 | 0.4984 | Yes |
| 9 | CLDN3 | CLDN3 Entrez, Source | claudin 3 | 504 | 0.181 | 0.5058 | Yes |
| 10 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 708 | 0.150 | 0.5121 | Yes |
| 11 | LMO4 | LMO4 Entrez, Source | LIM domain only 4 | 827 | 0.136 | 0.5230 | Yes |
| 12 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1068 | 0.116 | 0.5218 | Yes |
| 13 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 1250 | 0.107 | 0.5237 | Yes |
| 14 | ZFP36L1 | ZFP36L1 Entrez, Source | zinc finger protein 36, C3H type-like 1 | 1350 | 0.101 | 0.5309 | Yes |
| 15 | ACCN3 | ACCN3 Entrez, Source | amiloride-sensitive cation channel 3 | 1542 | 0.092 | 0.5298 | No |
| 16 | KCNIP4 | KCNIP4 Entrez, Source | Kv channel interacting protein 4 | 1705 | 0.085 | 0.5298 | No |
| 17 | CHRDL1 | CHRDL1 Entrez, Source | chordin-like 1 | 2416 | 0.062 | 0.4853 | No |
| 18 | CSPG4 | CSPG4 Entrez, Source | chondroitin sulfate proteoglycan 4 (melanoma-associated) | 2618 | 0.057 | 0.4784 | No |
| 19 | H3F3B | H3F3B Entrez, Source | H3 histone, family 3B (H3.3B) | 3095 | 0.047 | 0.4493 | No |
| 20 | VAMP3 | VAMP3 Entrez, Source | vesicle-associated membrane protein 3 (cellubrevin) | 3135 | 0.046 | 0.4530 | No |
| 21 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 3326 | 0.042 | 0.4447 | No |
| 22 | PHEX | PHEX Entrez, Source | phosphate regulating endopeptidase homolog, X-linked (hypophosphatemia, vitamin D resistant rickets) | 3518 | 0.038 | 0.4359 | No |
| 23 | ZFHX1B | ZFHX1B Entrez, Source | zinc finger homeobox 1b | 3802 | 0.033 | 0.4194 | No |
| 24 | DCX | DCX Entrez, Source | doublecortex; lissencephaly, X-linked (doublecortin) | 3837 | 0.033 | 0.4216 | No |
| 25 | PTGFR | PTGFR Entrez, Source | prostaglandin F receptor (FP) | 3885 | 0.032 | 0.4227 | No |
| 26 | ASAH1 | ASAH1 Entrez, Source | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 3904 | 0.032 | 0.4260 | No |
| 27 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 3967 | 0.031 | 0.4257 | No |
| 28 | PHYHIP | PHYHIP Entrez, Source | phytanoyl-CoA 2-hydroxylase interacting protein | 4233 | 0.026 | 0.4096 | No |
| 29 | APLN | APLN Entrez, Source | apelin, AGTRL1 ligand | 5207 | 0.013 | 0.3382 | No |
| 30 | RAB27A | RAB27A Entrez, Source | RAB27A, member RAS oncogene family | 5665 | 0.006 | 0.3046 | No |
| 31 | NRXN1 | NRXN1 Entrez, Source | neurexin 1 | 5683 | 0.006 | 0.3042 | No |
| 32 | FGF7 | FGF7 Entrez, Source | fibroblast growth factor 7 (keratinocyte growth factor) | 5875 | 0.004 | 0.2904 | No |
| 33 | RAP2B | RAP2B Entrez, Source | RAP2B, member of RAS oncogene family | 5893 | 0.004 | 0.2896 | No |
| 34 | ACVR1C | ACVR1C Entrez, Source | activin A receptor, type IC | 5910 | 0.003 | 0.2888 | No |
| 35 | AMPH | AMPH Entrez, Source | amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) | 5916 | 0.003 | 0.2889 | No |
| 36 | POFUT1 | POFUT1 Entrez, Source | protein O-fucosyltransferase 1 | 6279 | -0.002 | 0.2619 | No |
| 37 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 6398 | -0.003 | 0.2535 | No |
| 38 | ITGB3BP | ITGB3BP Entrez, Source | integrin beta 3 binding protein (beta3-endonexin) | 6849 | -0.009 | 0.2209 | No |
| 39 | CREM | CREM Entrez, Source | cAMP responsive element modulator | 6850 | -0.009 | 0.2221 | No |
| 40 | WNT16 | WNT16 Entrez, Source | wingless-type MMTV integration site family, member 16 | 7468 | -0.016 | 0.1780 | No |
| 41 | HTR2C | HTR2C Entrez, Source | 5-hydroxytryptamine (serotonin) receptor 2C | 7488 | -0.017 | 0.1790 | No |
| 42 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 7497 | -0.017 | 0.1808 | No |
| 43 | GATA3 | GATA3 Entrez, Source | GATA binding protein 3 | 7809 | -0.021 | 0.1604 | No |
| 44 | CITED1 | CITED1 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | 7841 | -0.021 | 0.1611 | No |
| 45 | DOCK11 | DOCK11 Entrez, Source | dedicator of cytokinesis 11 | 7970 | -0.023 | 0.1548 | No |
| 46 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 8552 | -0.031 | 0.1156 | No |
| 47 | OTX1 | OTX1 Entrez, Source | orthodenticle homolog 1 (Drosophila) | 8583 | -0.032 | 0.1180 | No |
| 48 | TPI1 | TPI1 Entrez, Source | triosephosphate isomerase 1 | 8794 | -0.035 | 0.1072 | No |
| 49 | HHEX | HHEX Entrez, Source | homeobox, hematopoietically expressed | 8800 | -0.035 | 0.1119 | No |
| 50 | HNRPC | HNRPC Entrez, Source | heterogeneous nuclear ribonucleoprotein C (C1/C2) | 9106 | -0.039 | 0.0946 | No |
| 51 | PPP2R2B | PPP2R2B Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform | 10110 | -0.055 | 0.0271 | No |
| 52 | CCKAR | CCKAR Entrez, Source | cholecystokinin A receptor | 10154 | -0.056 | 0.0319 | No |
| 53 | PCBP2 | PCBP2 Entrez, Source | poly(rC) binding protein 2 | 11317 | -0.082 | -0.0437 | No |
| 54 | NUDT4 | NUDT4 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 4 | 11563 | -0.088 | -0.0494 | No |
| 55 | RPS6KB1 | RPS6KB1 Entrez, Source | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | 11564 | -0.088 | -0.0367 | No |
| 56 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 12402 | -0.121 | -0.0821 | No |
| 57 | FGF17 | FGF17 Entrez, Source | fibroblast growth factor 17 | 12498 | -0.128 | -0.0708 | No |
| 58 | NUP54 | NUP54 Entrez, Source | nucleoporin 54kDa | 12635 | -0.137 | -0.0613 | No |
| 59 | BMF | BMF Entrez, Source | Bcl2 modifying factor | 12660 | -0.138 | -0.0431 | No |
| 60 | FAP | FAP Entrez, Source | fibroblast activation protein, alpha | 12774 | -0.149 | -0.0300 | No |
| 61 | LIMK2 | LIMK2 Entrez, Source | LIM domain kinase 2 | 13008 | -0.187 | -0.0205 | No |
| 62 | VLDLR | VLDLR Entrez, Source | very low density lipoprotein receptor | 13265 | -0.315 | 0.0058 | No |