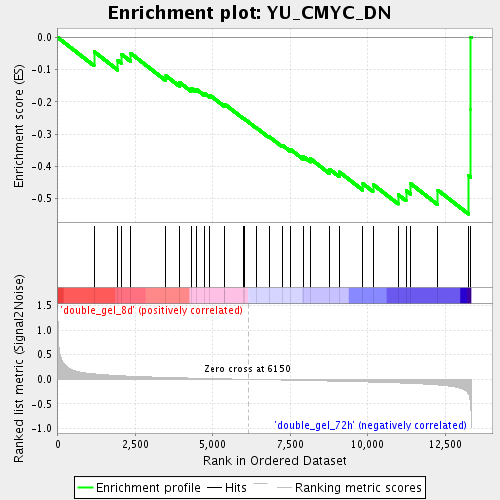
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_72h.class.cls #double_gel_8d_versus_double_gel_72h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | YU_CMYC_DN |
| Enrichment Score (ES) | -0.54913217 |
| Normalized Enrichment Score (NES) | -1.6939319 |
| Nominal p-value | 0.0069124424 |
| FDR q-value | 0.16179435 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RGL2 | RGL2 Entrez, Source | ral guanine nucleotide dissociation stimulator-like 2 | 1167 | 0.111 | -0.0436 | No |
| 2 | RBMS1 | RBMS1 Entrez, Source | RNA binding motif, single stranded interacting protein 1 | 1934 | 0.075 | -0.0713 | No |
| 3 | PTPRC | PTPRC Entrez, Source | protein tyrosine phosphatase, receptor type, C | 2044 | 0.071 | -0.0512 | No |
| 4 | BTG1 | BTG1 Entrez, Source | B-cell translocation gene 1, anti-proliferative | 2350 | 0.063 | -0.0490 | No |
| 5 | GDI1 | GDI1 Entrez, Source | GDP dissociation inhibitor 1 | 3486 | 0.039 | -0.1188 | No |
| 6 | DGKA | DGKA Entrez, Source | diacylglycerol kinase, alpha 80kDa | 3925 | 0.031 | -0.1393 | No |
| 7 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 4298 | 0.025 | -0.1571 | No |
| 8 | MGST1 | MGST1 Entrez, Source | microsomal glutathione S-transferase 1 | 4459 | 0.023 | -0.1601 | No |
| 9 | CD2 | CD2 Entrez, Source | CD2 molecule | 4735 | 0.019 | -0.1731 | No |
| 10 | OLFM1 | OLFM1 Entrez, Source | olfactomedin 1 | 4900 | 0.017 | -0.1788 | No |
| 11 | SRPK3 | SRPK3 Entrez, Source | SFRS protein kinase 3 | 5371 | 0.010 | -0.2101 | No |
| 12 | CTSH | CTSH Entrez, Source | cathepsin H | 5387 | 0.010 | -0.2073 | No |
| 13 | CMAH | CMAH Entrez, Source | cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) | 5991 | 0.002 | -0.2517 | No |
| 14 | IL10RA | IL10RA Entrez, Source | interleukin 10 receptor, alpha | 6007 | 0.002 | -0.2520 | No |
| 15 | EVL | EVL Entrez, Source | Enah/Vasp-like | 6399 | -0.003 | -0.2801 | No |
| 16 | IGH-1A | 6813 | -0.008 | -0.3078 | No | ||
| 17 | LAPTM5 | LAPTM5 Entrez, Source | lysosomal associated multispanning membrane protein 5 | 7239 | -0.014 | -0.3344 | No |
| 18 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 7497 | -0.017 | -0.3471 | No |
| 19 | UNC93B1 | UNC93B1 Entrez, Source | unc-93 homolog B1 (C. elegans) | 7931 | -0.022 | -0.3707 | No |
| 20 | BLK | BLK Entrez, Source | B lymphoid tyrosine kinase | 8151 | -0.026 | -0.3769 | No |
| 21 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 8764 | -0.034 | -0.4093 | No |
| 22 | ARHGEF1 | ARHGEF1 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 1 | 9085 | -0.039 | -0.4180 | No |
| 23 | GGA2 | GGA2 Entrez, Source | golgi associated, gamma adaptin ear containing, ARF binding protein 2 | 9841 | -0.051 | -0.4545 | No |
| 24 | CD37 | CD37 Entrez, Source | CD37 molecule | 10171 | -0.056 | -0.4569 | No |
| 25 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 10982 | -0.073 | -0.4888 | Yes |
| 26 | CD1D1 | 11237 | -0.079 | -0.4764 | Yes | ||
| 27 | ACP5 | ACP5 Entrez, Source | acid phosphatase 5, tartrate resistant | 11379 | -0.083 | -0.4539 | Yes |
| 28 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 12263 | -0.115 | -0.4748 | Yes |
| 29 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 13254 | -0.302 | -0.4292 | Yes |
| 30 | CD74 | CD74 Entrez, Source | CD74 molecule, major histocompatibility complex, class II invariant chain | 13324 | -0.530 | -0.2241 | Yes |
| 31 | LTB | LTB Entrez, Source | lymphotoxin beta (TNF superfamily, member 3) | 13327 | -0.568 | 0.0011 | Yes |

