Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
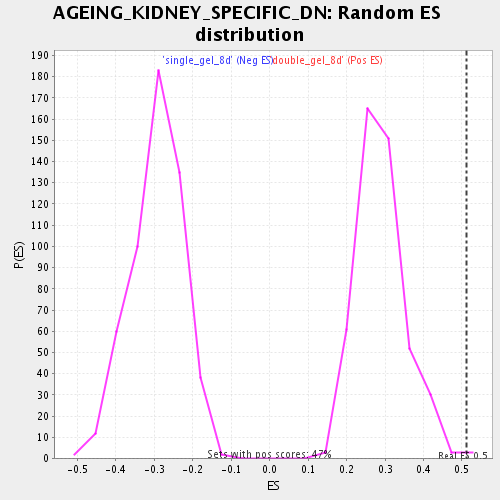
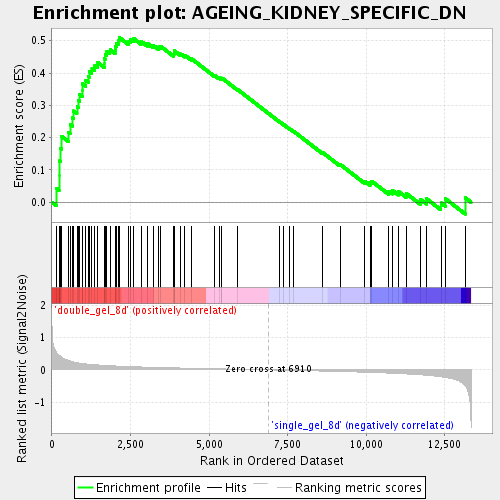
| GeneSet | AGEING_KIDNEY_SPECIFIC_DN |
| Enrichment Score (ES) | 0.51070875 |
| Normalized Enrichment Score (NES) | 1.7639769 |
| Nominal p-value | 0.0042735045 |
| FDR q-value | 0.019317938 |
| FWER p-Value | 0.962 |

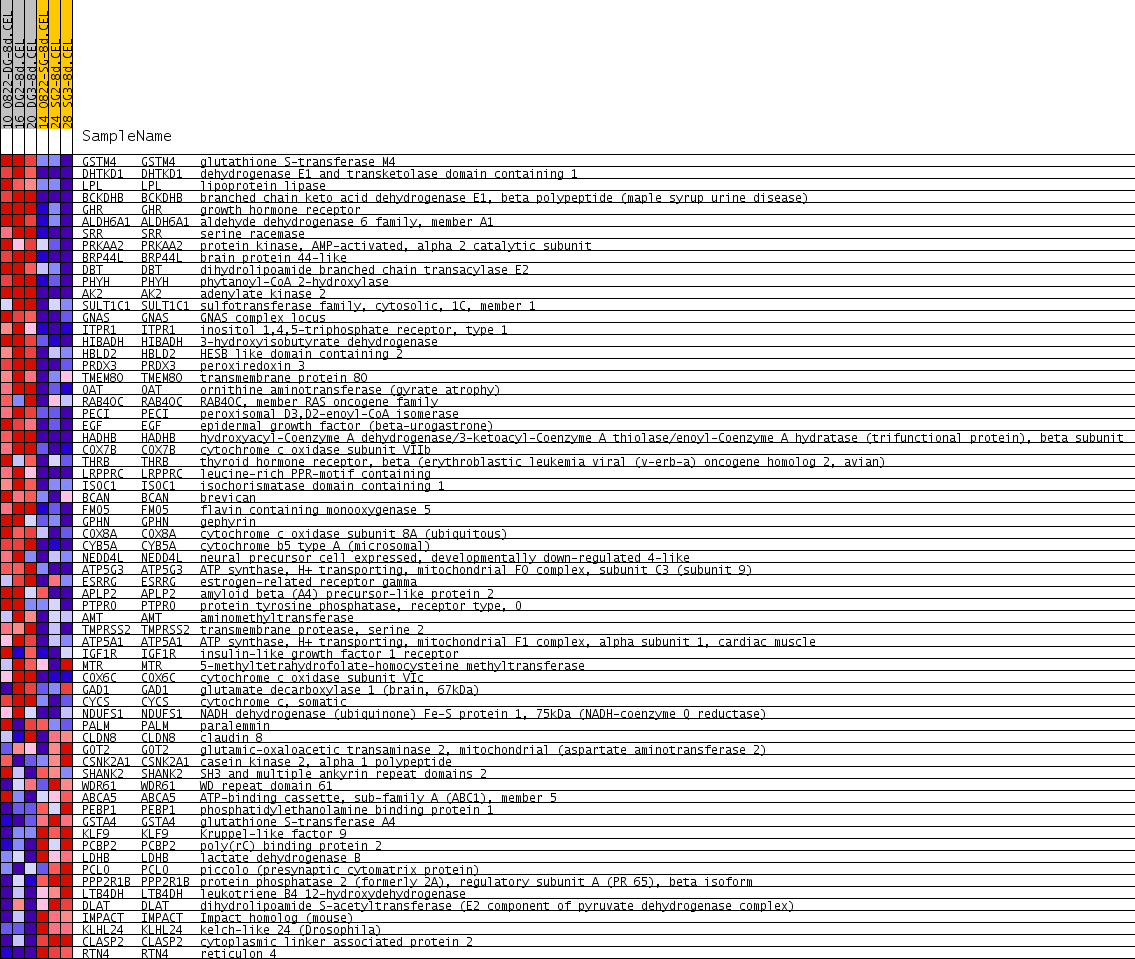
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GSTM4 | GSTM4 Entrez, Source | glutathione S-transferase M4 | 151 | 0.530 | 0.0422 | Yes |
| 2 | DHTKD1 | DHTKD1 Entrez, Source | dehydrogenase E1 and transketolase domain containing 1 | 228 | 0.454 | 0.0824 | Yes |
| 3 | LPL | LPL Entrez, Source | lipoprotein lipase | 230 | 0.451 | 0.1279 | Yes |
| 4 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 271 | 0.419 | 0.1673 | Yes |
| 5 | GHR | GHR Entrez, Source | growth hormone receptor | 312 | 0.389 | 0.2036 | Yes |
| 6 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 524 | 0.292 | 0.2172 | Yes |
| 7 | SRR | SRR Entrez, Source | serine racemase | 579 | 0.274 | 0.2408 | Yes |
| 8 | PRKAA2 | PRKAA2 Entrez, Source | protein kinase, AMP-activated, alpha 2 catalytic subunit | 649 | 0.252 | 0.2611 | Yes |
| 9 | BRP44L | BRP44L Entrez, Source | brain protein 44-like | 688 | 0.241 | 0.2826 | Yes |
| 10 | DBT | DBT Entrez, Source | dihydrolipoamide branched chain transacylase E2 | 799 | 0.220 | 0.2966 | Yes |
| 11 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 849 | 0.214 | 0.3145 | Yes |
| 12 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 880 | 0.210 | 0.3335 | Yes |
| 13 | SULT1C1 | SULT1C1 Entrez, Source | sulfotransferase family, cytosolic, 1C, member 1 | 964 | 0.199 | 0.3473 | Yes |
| 14 | GNAS | GNAS Entrez, Source | GNAS complex locus | 968 | 0.198 | 0.3671 | Yes |
| 15 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 1073 | 0.185 | 0.3779 | Yes |
| 16 | HIBADH | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 1157 | 0.175 | 0.3894 | Yes |
| 17 | HBLD2 | HBLD2 Entrez, Source | HESB like domain containing 2 | 1186 | 0.172 | 0.4046 | Yes |
| 18 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 1274 | 0.166 | 0.4148 | Yes |
| 19 | TMEM80 | TMEM80 Entrez, Source | transmembrane protein 80 | 1365 | 0.160 | 0.4242 | Yes |
| 20 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 1460 | 0.152 | 0.4325 | Yes |
| 21 | RAB40C | RAB40C Entrez, Source | RAB40C, member RAS oncogene family | 1677 | 0.138 | 0.4302 | Yes |
| 22 | PECI | PECI Entrez, Source | peroxisomal D3,D2-enoyl-CoA isomerase | 1680 | 0.138 | 0.4440 | Yes |
| 23 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1695 | 0.137 | 0.4568 | Yes |
| 24 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1747 | 0.134 | 0.4665 | Yes |
| 25 | COX7B | COX7B Entrez, Source | cytochrome c oxidase subunit VIIb | 1849 | 0.129 | 0.4719 | Yes |
| 26 | THRB | THRB Entrez, Source | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | 2013 | 0.121 | 0.4719 | Yes |
| 27 | LRPPRC | LRPPRC Entrez, Source | leucine-rich PPR-motif containing | 2030 | 0.121 | 0.4829 | Yes |
| 28 | ISOC1 | ISOC1 Entrez, Source | isochorismatase domain containing 1 | 2061 | 0.119 | 0.4926 | Yes |
| 29 | BCAN | BCAN Entrez, Source | brevican | 2126 | 0.117 | 0.4996 | Yes |
| 30 | FMO5 | FMO5 Entrez, Source | flavin containing monooxygenase 5 | 2136 | 0.117 | 0.5107 | Yes |
| 31 | GPHN | GPHN Entrez, Source | gephyrin | 2447 | 0.105 | 0.4979 | No |
| 32 | COX8A | COX8A Entrez, Source | cytochrome c oxidase subunit 8A (ubiquitous) | 2501 | 0.102 | 0.5043 | No |
| 33 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 2608 | 0.099 | 0.5063 | No |
| 34 | NEDD4L | NEDD4L Entrez, Source | neural precursor cell expressed, developmentally down-regulated 4-like | 2839 | 0.090 | 0.4981 | No |
| 35 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 3038 | 0.084 | 0.4916 | No |
| 36 | ESRRG | ESRRG Entrez, Source | estrogen-related receptor gamma | 3222 | 0.078 | 0.4857 | No |
| 37 | APLP2 | APLP2 Entrez, Source | amyloid beta (A4) precursor-like protein 2 | 3389 | 0.074 | 0.4807 | No |
| 38 | PTPRO | PTPRO Entrez, Source | protein tyrosine phosphatase, receptor type, O | 3457 | 0.072 | 0.4829 | No |
| 39 | AMT | AMT Entrez, Source | aminomethyltransferase | 3875 | 0.063 | 0.4578 | No |
| 40 | TMPRSS2 | TMPRSS2 Entrez, Source | transmembrane protease, serine 2 | 3889 | 0.062 | 0.4631 | No |
| 41 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 3895 | 0.062 | 0.4690 | No |
| 42 | IGF1R | IGF1R Entrez, Source | insulin-like growth factor 1 receptor | 4105 | 0.057 | 0.4591 | No |
| 43 | MTR | MTR Entrez, Source | 5-methyltetrahydrofolate-homocysteine methyltransferase | 4234 | 0.054 | 0.4549 | No |
| 44 | COX6C | COX6C Entrez, Source | cytochrome c oxidase subunit VIc | 4441 | 0.050 | 0.4444 | No |
| 45 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5183 | 0.034 | 0.3921 | No |
| 46 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 5320 | 0.032 | 0.3850 | No |
| 47 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5395 | 0.030 | 0.3825 | No |
| 48 | PALM | PALM Entrez, Source | paralemmin | 5408 | 0.030 | 0.3846 | No |
| 49 | CLDN8 | CLDN8 Entrez, Source | claudin 8 | 5910 | 0.020 | 0.3489 | No |
| 50 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 7234 | -0.007 | 0.2500 | No |
| 51 | CSNK2A1 | CSNK2A1 Entrez, Source | casein kinase 2, alpha 1 polypeptide | 7384 | -0.010 | 0.2398 | No |
| 52 | SHANK2 | SHANK2 Entrez, Source | SH3 and multiple ankyrin repeat domains 2 | 7570 | -0.014 | 0.2272 | No |
| 53 | WDR61 | WDR61 Entrez, Source | WD repeat domain 61 | 7683 | -0.016 | 0.2204 | No |
| 54 | ABCA5 | ABCA5 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 5 | 8600 | -0.037 | 0.1552 | No |
| 55 | PEBP1 | PEBP1 Entrez, Source | phosphatidylethanolamine binding protein 1 | 9172 | -0.051 | 0.1173 | No |
| 56 | GSTA4 | GSTA4 Entrez, Source | glutathione S-transferase A4 | 9961 | -0.073 | 0.0653 | No |
| 57 | KLF9 | KLF9 Entrez, Source | Kruppel-like factor 9 | 10129 | -0.079 | 0.0607 | No |
| 58 | PCBP2 | PCBP2 Entrez, Source | poly(rC) binding protein 2 | 10175 | -0.081 | 0.0655 | No |
| 59 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 10723 | -0.099 | 0.0343 | No |
| 60 | PCLO | PCLO Entrez, Source | piccolo (presynaptic cytomatrix protein) | 10844 | -0.105 | 0.0359 | No |
| 61 | PPP2R1B | PPP2R1B Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform | 11027 | -0.113 | 0.0336 | No |
| 62 | LTB4DH | LTB4DH Entrez, Source | leukotriene B4 12-hydroxydehydrogenase | 11276 | -0.125 | 0.0276 | No |
| 63 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 11723 | -0.150 | 0.0092 | No |
| 64 | IMPACT | IMPACT Entrez, Source | Impact homolog (mouse) | 11924 | -0.167 | 0.0110 | No |
| 65 | KLHL24 | KLHL24 Entrez, Source | kelch-like 24 (Drosophila) | 12386 | -0.213 | -0.0022 | No |
| 66 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 12533 | -0.233 | 0.0104 | No |
| 67 | RTN4 | RTN4 Entrez, Source | reticulon 4 | 13160 | -0.500 | 0.0137 | No |