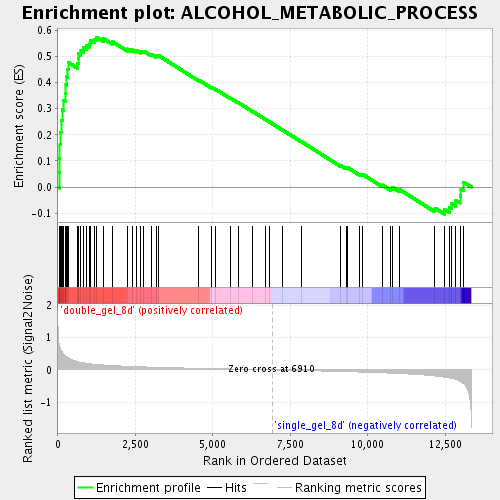
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | ALCOHOL_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5737953 |
| Normalized Enrichment Score (NES) | 1.9230065 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004225609 |
| FWER p-Value | 0.351 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 50 | 0.751 | 0.0544 | Yes |
| 2 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 58 | 0.726 | 0.1100 | Yes |
| 3 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 66 | 0.702 | 0.1638 | Yes |
| 4 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 95 | 0.621 | 0.2098 | Yes |
| 5 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 103 | 0.612 | 0.2566 | Yes |
| 6 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 143 | 0.545 | 0.2959 | Yes |
| 7 | NR0B2 | NR0B2 Entrez, Source | nuclear receptor subfamily 0, group B, member 2 | 187 | 0.486 | 0.3303 | Yes |
| 8 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 247 | 0.438 | 0.3597 | Yes |
| 9 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.3920 | Yes |
| 10 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 282 | 0.412 | 0.4222 | Yes |
| 11 | FDPS | FDPS Entrez, Source | farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | 315 | 0.386 | 0.4497 | Yes |
| 12 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 333 | 0.375 | 0.4774 | Yes |
| 13 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 642 | 0.255 | 0.4740 | Yes |
| 14 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 656 | 0.251 | 0.4925 | Yes |
| 15 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 666 | 0.249 | 0.5111 | Yes |
| 16 | SULT1B1 | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 743 | 0.229 | 0.5231 | Yes |
| 17 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 829 | 0.216 | 0.5334 | Yes |
| 18 | COQ3 | COQ3 Entrez, Source | coenzyme Q3 homolog, methyltransferase (S. cerevisiae) | 932 | 0.202 | 0.5414 | Yes |
| 19 | GALK1 | GALK1 Entrez, Source | galactokinase 1 | 1032 | 0.189 | 0.5486 | Yes |
| 20 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 1047 | 0.187 | 0.5620 | Yes |
| 21 | CLN8 | CLN8 Entrez, Source | ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) | 1178 | 0.173 | 0.5656 | Yes |
| 22 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 1243 | 0.168 | 0.5738 | Yes |
| 23 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 1462 | 0.152 | 0.5692 | No |
| 24 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1770 | 0.133 | 0.5563 | No |
| 25 | SCARB1 | SCARB1 Entrez, Source | scavenger receptor class B, member 1 | 2252 | 0.112 | 0.5288 | No |
| 26 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 2402 | 0.107 | 0.5258 | No |
| 27 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 2527 | 0.102 | 0.5244 | No |
| 28 | CEL | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 2678 | 0.096 | 0.5206 | No |
| 29 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 2767 | 0.093 | 0.5211 | No |
| 30 | ARPP-19 | ARPP-19 Entrez, Source | - | 3013 | 0.084 | 0.5092 | No |
| 31 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 3168 | 0.079 | 0.5038 | No |
| 32 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 3231 | 0.078 | 0.5051 | No |
| 33 | IL4 | IL4 Entrez, Source | interleukin 4 | 4550 | 0.047 | 0.4096 | No |
| 34 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 4971 | 0.038 | 0.3809 | No |
| 35 | HDLBP | HDLBP Entrez, Source | high density lipoprotein binding protein (vigilin) | 5095 | 0.036 | 0.3745 | No |
| 36 | IPPK | IPPK Entrez, Source | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 5568 | 0.026 | 0.3410 | No |
| 37 | ADH4 | ADH4 Entrez, Source | alcohol dehydrogenase 4 (class II), pi polypeptide | 5822 | 0.022 | 0.3236 | No |
| 38 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.2902 | No |
| 39 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 6698 | 0.004 | 0.2591 | No |
| 40 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 6814 | 0.002 | 0.2506 | No |
| 41 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 7238 | -0.007 | 0.2193 | No |
| 42 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7856 | -0.020 | 0.1744 | No |
| 43 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 9121 | -0.049 | 0.0830 | No |
| 44 | CNBP | CNBP Entrez, Source | CCHC-type zinc finger, nucleic acid binding protein | 9302 | -0.054 | 0.0737 | No |
| 45 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 9345 | -0.056 | 0.0748 | No |
| 46 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9745 | -0.066 | 0.0499 | No |
| 47 | GALT | GALT Entrez, Source | galactose-1-phosphate uridylyltransferase | 9817 | -0.069 | 0.0499 | No |
| 48 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10469 | -0.091 | 0.0079 | No |
| 49 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 10743 | -0.100 | -0.0049 | No |
| 50 | MBTPS2 | MBTPS2 Entrez, Source | membrane-bound transcription factor peptidase, site 2 | 10788 | -0.102 | -0.0003 | No |
| 51 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 11033 | -0.113 | -0.0098 | No |
| 52 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 12169 | -0.189 | -0.0807 | No |
| 53 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 12474 | -0.224 | -0.0862 | No |
| 54 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12636 | -0.251 | -0.0789 | No |
| 55 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12700 | -0.264 | -0.0632 | No |
| 56 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12832 | -0.296 | -0.0502 | No |
| 57 | SOAT1 | SOAT1 Entrez, Source | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 12979 | -0.351 | -0.0340 | No |
| 58 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 13007 | -0.369 | -0.0075 | No |
| 59 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 13094 | -0.421 | 0.0187 | No |

