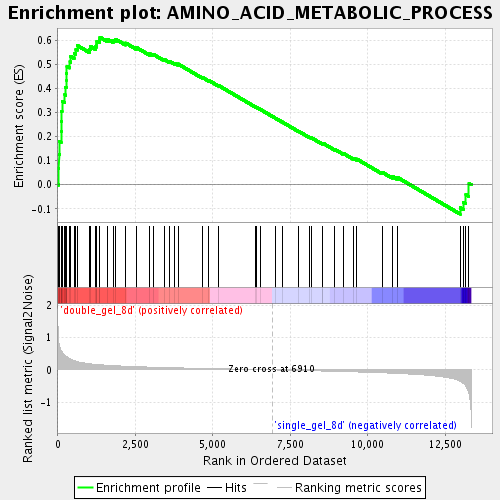
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | AMINO_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.61264104 |
| Normalized Enrichment Score (NES) | 2.0546854 |
| Nominal p-value | 0.0 |
| FDR q-value | 9.7100885E-4 |
| FWER p-Value | 0.066 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 17 | 0.930 | 0.0673 | Yes |
| 2 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 35 | 0.815 | 0.1261 | Yes |
| 3 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 56 | 0.731 | 0.1785 | Yes |
| 4 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 104 | 0.612 | 0.2201 | Yes |
| 5 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 113 | 0.597 | 0.2635 | Yes |
| 6 | ASL | ASL Entrez, Source | argininosuccinate lyase | 122 | 0.586 | 0.3061 | Yes |
| 7 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 135 | 0.556 | 0.3463 | Yes |
| 8 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 223 | 0.456 | 0.3733 | Yes |
| 9 | ARG1 | ARG1 Entrez, Source | arginase, liver | 246 | 0.438 | 0.4040 | Yes |
| 10 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 271 | 0.419 | 0.4331 | Yes |
| 11 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 277 | 0.416 | 0.4634 | Yes |
| 12 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 291 | 0.403 | 0.4922 | Yes |
| 13 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 389 | 0.344 | 0.5103 | Yes |
| 14 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 411 | 0.336 | 0.5335 | Yes |
| 15 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 524 | 0.292 | 0.5466 | Yes |
| 16 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 580 | 0.273 | 0.5626 | Yes |
| 17 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 623 | 0.260 | 0.5786 | Yes |
| 18 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 1027 | 0.189 | 0.5623 | Yes |
| 19 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 1046 | 0.187 | 0.5747 | Yes |
| 20 | SDS | SDS Entrez, Source | serine dehydratase | 1222 | 0.169 | 0.5740 | Yes |
| 21 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 1252 | 0.167 | 0.5841 | Yes |
| 22 | HDC | HDC Entrez, Source | histidine decarboxylase | 1256 | 0.167 | 0.5962 | Yes |
| 23 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 1345 | 0.161 | 0.6015 | Yes |
| 24 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 1355 | 0.160 | 0.6126 | Yes |
| 25 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 1598 | 0.143 | 0.6050 | No |
| 26 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 1785 | 0.132 | 0.6007 | No |
| 27 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 1864 | 0.128 | 0.6043 | No |
| 28 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2183 | 0.115 | 0.5888 | No |
| 29 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 2527 | 0.102 | 0.5705 | No |
| 30 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 2968 | 0.086 | 0.5437 | No |
| 31 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 3082 | 0.082 | 0.5413 | No |
| 32 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 3436 | 0.073 | 0.5201 | No |
| 33 | GSS | GSS Entrez, Source | glutathione synthetase | 3596 | 0.069 | 0.5132 | No |
| 34 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 3774 | 0.065 | 0.5046 | No |
| 35 | AMT | AMT Entrez, Source | aminomethyltransferase | 3875 | 0.063 | 0.5017 | No |
| 36 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 4665 | 0.045 | 0.4456 | No |
| 37 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 4874 | 0.040 | 0.4329 | No |
| 38 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5183 | 0.034 | 0.4123 | No |
| 39 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 6381 | 0.010 | 0.3229 | No |
| 40 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 6420 | 0.009 | 0.3207 | No |
| 41 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 6525 | 0.007 | 0.3135 | No |
| 42 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 7015 | -0.002 | 0.2768 | No |
| 43 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 7234 | -0.007 | 0.2609 | No |
| 44 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 7757 | -0.018 | 0.2230 | No |
| 45 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 8124 | -0.026 | 0.1973 | No |
| 46 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 8177 | -0.027 | 0.1954 | No |
| 47 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 8542 | -0.036 | 0.1706 | No |
| 48 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 8548 | -0.036 | 0.1729 | No |
| 49 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 8939 | -0.045 | 0.1468 | No |
| 50 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 9206 | -0.051 | 0.1306 | No |
| 51 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 9551 | -0.061 | 0.1092 | No |
| 52 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 9642 | -0.064 | 0.1071 | No |
| 53 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 10462 | -0.090 | 0.0521 | No |
| 54 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 10791 | -0.102 | 0.0350 | No |
| 55 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 10959 | -0.110 | 0.0305 | No |
| 56 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 12983 | -0.354 | -0.0957 | No |
| 57 | SMS | SMS Entrez, Source | spermine synthase | 13083 | -0.414 | -0.0725 | No |
| 58 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13167 | -0.505 | -0.0416 | No |
| 59 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 13266 | -0.741 | 0.0057 | No |