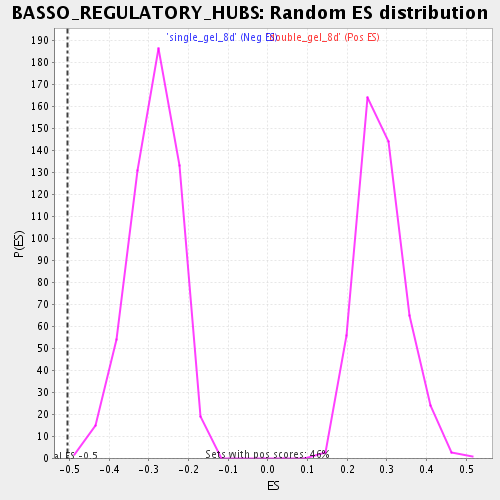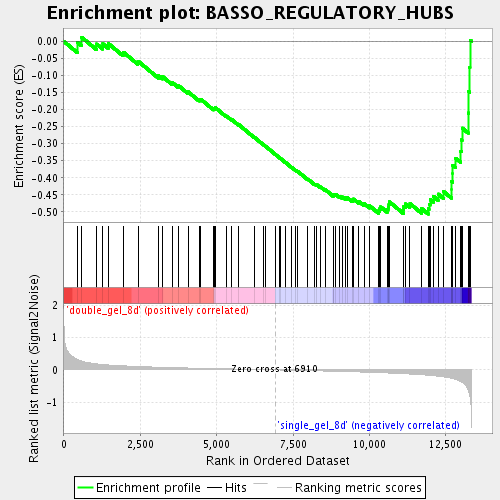
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | BASSO_REGULATORY_HUBS |
| Enrichment Score (ES) | -0.5064539 |
| Normalized Enrichment Score (NES) | -1.749942 |
| Nominal p-value | 0.0018518518 |
| FDR q-value | 0.063532285 |
| FWER p-Value | 0.993 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LGALS9 | LGALS9 Entrez, Source | lectin, galactoside-binding, soluble, 9 (galectin 9) | 440 | 0.324 | -0.0028 | No |
| 2 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 581 | 0.272 | 0.0122 | No |
| 3 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 1073 | 0.185 | -0.0075 | No |
| 4 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 1274 | 0.166 | -0.0071 | No |
| 5 | AFG3L2 | AFG3L2 Entrez, Source | AFG3 ATPase family gene 3-like 2 (yeast) | 1449 | 0.153 | -0.0059 | No |
| 6 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 1940 | 0.124 | -0.0311 | No |
| 7 | ASNS | ASNS Entrez, Source | asparagine synthetase | 2424 | 0.106 | -0.0576 | No |
| 8 | CASP4 | CASP4 Entrez, Source | caspase 4, apoptosis-related cysteine peptidase | 3096 | 0.082 | -0.1006 | No |
| 9 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 3227 | 0.078 | -0.1031 | No |
| 10 | TGOLN2 | TGOLN2 Entrez, Source | trans-golgi network protein 2 | 3548 | 0.070 | -0.1206 | No |
| 11 | LAPTM5 | LAPTM5 Entrez, Source | lysosomal associated multispanning membrane protein 5 | 3735 | 0.066 | -0.1285 | No |
| 12 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 4062 | 0.058 | -0.1477 | No |
| 13 | PSMD1 | PSMD1 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 4425 | 0.050 | -0.1703 | No |
| 14 | BUD31 | BUD31 Entrez, Source | BUD31 homolog (yeast) | 4471 | 0.049 | -0.1691 | No |
| 15 | PRG1 | PRG1 Entrez, Source | proteoglycan 1, secretory granule | 4898 | 0.040 | -0.1974 | No |
| 16 | PLCB2 | PLCB2 Entrez, Source | phospholipase C, beta 2 | 4924 | 0.039 | -0.1956 | No |
| 17 | LITAF | LITAF Entrez, Source | lipopolysaccharide-induced TNF factor | 4957 | 0.039 | -0.1944 | No |
| 18 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 5320 | 0.032 | -0.2187 | No |
| 19 | GARS | GARS Entrez, Source | glycyl-tRNA synthetase | 5484 | 0.028 | -0.2284 | No |
| 20 | TCEB1 | TCEB1 Entrez, Source | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | 5725 | 0.023 | -0.2443 | No |
| 21 | EPOR | EPOR Entrez, Source | erythropoietin receptor | 6228 | 0.014 | -0.2809 | No |
| 22 | PSMB2 | PSMB2 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 2 | 6524 | 0.007 | -0.3024 | No |
| 23 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 6598 | 0.006 | -0.3074 | No |
| 24 | PSMB7 | PSMB7 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 7 | 6920 | -0.000 | -0.3315 | No |
| 25 | SSBP1 | SSBP1 Entrez, Source | single-stranded DNA binding protein 1 | 7057 | -0.003 | -0.3415 | No |
| 26 | GABARAP | GABARAP Entrez, Source | GABA(A) receptor-associated protein | 7091 | -0.004 | -0.3436 | No |
| 27 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 7238 | -0.007 | -0.3540 | No |
| 28 | MVP | MVP Entrez, Source | major vault protein | 7452 | -0.011 | -0.3690 | No |
| 29 | SAR1A | SAR1A Entrez, Source | SAR1 gene homolog A (S. cerevisiae) | 7581 | -0.014 | -0.3773 | No |
| 30 | BYSL | BYSL Entrez, Source | bystin-like | 7639 | -0.015 | -0.3801 | No |
| 31 | SLC4A3 | SLC4A3 Entrez, Source | solute carrier family 4, anion exchanger, member 3 | 7978 | -0.023 | -0.4035 | No |
| 32 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 8212 | -0.028 | -0.4184 | No |
| 33 | HNRPAB | HNRPAB Entrez, Source | heterogeneous nuclear ribonucleoprotein A/B | 8280 | -0.029 | -0.4207 | No |
| 34 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 8399 | -0.032 | -0.4266 | No |
| 35 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 8548 | -0.036 | -0.4344 | No |
| 36 | ZMYND11 | ZMYND11 Entrez, Source | zinc finger, MYND domain containing 11 | 8813 | -0.041 | -0.4504 | No |
| 37 | RPS26 | RPS26 Entrez, Source | ribosomal protein S26 | 8833 | -0.042 | -0.4480 | No |
| 38 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 8885 | -0.043 | -0.4477 | No |
| 39 | ITPR2 | ITPR2 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 2 | 9021 | -0.046 | -0.4536 | No |
| 40 | SERBP1 | SERBP1 Entrez, Source | SERPINE1 mRNA binding protein 1 | 9117 | -0.049 | -0.4561 | No |
| 41 | PSMB5 | PSMB5 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 5 | 9216 | -0.052 | -0.4587 | No |
| 42 | PSMD2 | PSMD2 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 9272 | -0.054 | -0.4578 | No |
| 43 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 9446 | -0.058 | -0.4654 | No |
| 44 | TEP1 | TEP1 Entrez, Source | telomerase-associated protein 1 | 9476 | -0.059 | -0.4620 | No |
| 45 | WDR18 | WDR18 Entrez, Source | WD repeat domain 18 | 9648 | -0.064 | -0.4689 | No |
| 46 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 9827 | -0.069 | -0.4759 | No |
| 47 | EIF2S1 | EIF2S1 Entrez, Source | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | 9999 | -0.075 | -0.4818 | No |
| 48 | TCP1 | TCP1 Entrez, Source | t-complex 1 | 10306 | -0.085 | -0.4969 | Yes |
| 49 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 10338 | -0.086 | -0.4912 | Yes |
| 50 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 10369 | -0.087 | -0.4853 | Yes |
| 51 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 10578 | -0.094 | -0.4921 | Yes |
| 52 | MGEA5 | MGEA5 Entrez, Source | meningioma expressed antigen 5 (hyaluronidase) | 10610 | -0.095 | -0.4855 | Yes |
| 53 | SSRP1 | SSRP1 Entrez, Source | structure specific recognition protein 1 | 10636 | -0.096 | -0.4784 | Yes |
| 54 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 10651 | -0.097 | -0.4703 | Yes |
| 55 | BLMH | BLMH Entrez, Source | bleomycin hydrolase | 11109 | -0.117 | -0.4938 | Yes |
| 56 | HHEX | HHEX Entrez, Source | homeobox, hematopoietically expressed | 11122 | -0.118 | -0.4836 | Yes |
| 57 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 11178 | -0.120 | -0.4765 | Yes |
| 58 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 11324 | -0.128 | -0.4755 | Yes |
| 59 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 11707 | -0.150 | -0.4902 | Yes |
| 60 | RBM5 | RBM5 Entrez, Source | RNA binding motif protein 5 | 11923 | -0.167 | -0.4908 | Yes |
| 61 | CTSH | CTSH Entrez, Source | cathepsin H | 11951 | -0.169 | -0.4770 | Yes |
| 62 | TPP1 | TPP1 Entrez, Source | tripeptidyl peptidase I | 11989 | -0.172 | -0.4637 | Yes |
| 63 | LGALS1 | LGALS1 Entrez, Source | lectin, galactoside-binding, soluble, 1 (galectin 1) | 12094 | -0.181 | -0.4545 | Yes |
| 64 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 12248 | -0.199 | -0.4475 | Yes |
| 65 | BIN1 | BIN1 Entrez, Source | bridging integrator 1 | 12429 | -0.219 | -0.4405 | Yes |
| 66 | NISCH | NISCH Entrez, Source | nischarin | 12686 | -0.261 | -0.4353 | Yes |
| 67 | S100A4 | S100A4 Entrez, Source | S100 calcium binding protein A4 | 12689 | -0.262 | -0.4110 | Yes |
| 68 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 12720 | -0.269 | -0.3880 | Yes |
| 69 | ZYX | ZYX Entrez, Source | zyxin | 12724 | -0.270 | -0.3629 | Yes |
| 70 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 12822 | -0.294 | -0.3427 | Yes |
| 71 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 12986 | -0.357 | -0.3215 | Yes |
| 72 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13016 | -0.374 | -0.2886 | Yes |
| 73 | UCHL1 | UCHL1 Entrez, Source | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 13032 | -0.382 | -0.2540 | Yes |
| 74 | CD44 | CD44 Entrez, Source | CD44 molecule (Indian blood group) | 13233 | -0.633 | -0.2098 | Yes |
| 75 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13247 | -0.677 | -0.1473 | Yes |
| 76 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 13282 | -0.795 | -0.0753 | Yes |
| 77 | AHNAK | AHNAK Entrez, Source | AHNAK nucleoprotein (desmoyokin) | 13291 | -0.852 | 0.0038 | Yes |