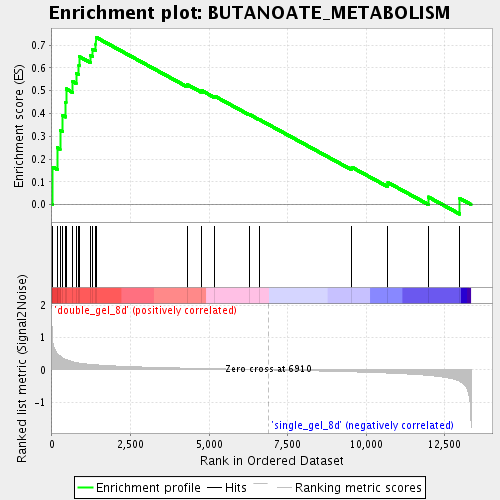
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | BUTANOATE_METABOLISM |
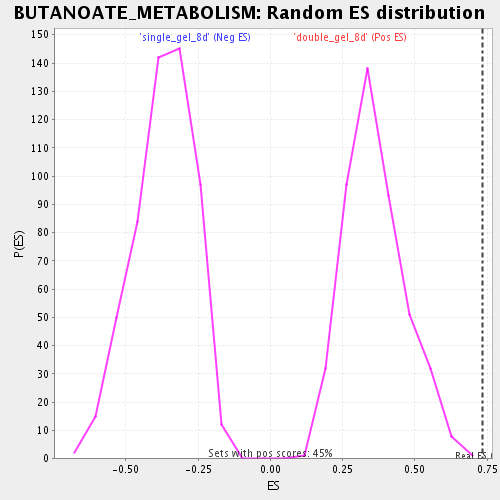
| Enrichment Score (ES) | 0.7347597 |
| Normalized Enrichment Score (NES) | 2.0214517 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0012391097 |
| FWER p-Value | 0.098 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 24 | 0.864 | 0.1647 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 172 | 0.499 | 0.2498 | Yes |
| 3 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.3255 | Yes |
| 4 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 344 | 0.371 | 0.3907 | Yes |
| 5 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 430 | 0.329 | 0.4478 | Yes |
| 6 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 450 | 0.320 | 0.5081 | Yes |
| 7 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 663 | 0.250 | 0.5402 | Yes |
| 8 | AACS | AACS Entrez, Source | acetoacetyl-CoA synthetase | 771 | 0.224 | 0.5754 | Yes |
| 9 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.6100 | Yes |
| 10 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 870 | 0.211 | 0.6499 | Yes |
| 11 | SDS | SDS Entrez, Source | serine dehydratase | 1222 | 0.169 | 0.6560 | Yes |
| 12 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1296 | 0.164 | 0.6822 | Yes |
| 13 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1399 | 0.157 | 0.7048 | Yes |
| 14 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 1403 | 0.157 | 0.7348 | Yes |
| 15 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 4305 | 0.053 | 0.5271 | No |
| 16 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 4772 | 0.042 | 0.5003 | No |
| 17 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5183 | 0.034 | 0.4761 | No |
| 18 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.3963 | No |
| 19 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 6602 | 0.006 | 0.3733 | No |
| 20 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 9551 | -0.061 | 0.1637 | No |
| 21 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 10696 | -0.098 | 0.0968 | No |
| 22 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 11983 | -0.171 | 0.0332 | No |
| 23 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 12985 | -0.357 | 0.0268 | No |