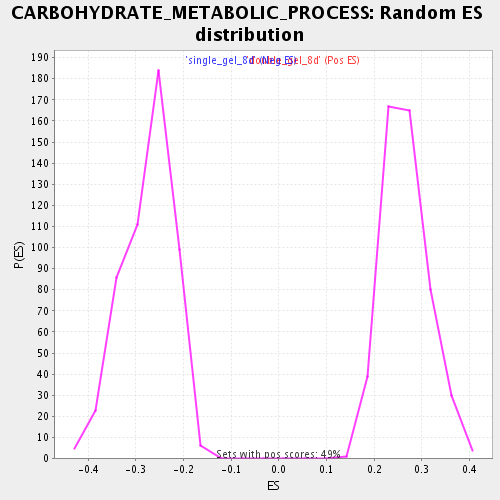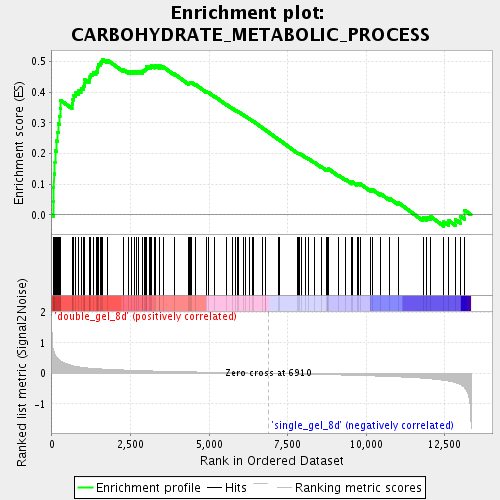
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CARBOHYDRATE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5077968 |
| Normalized Enrichment Score (NES) | 1.9119837 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004745326 |
| FWER p-Value | 0.4 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GYS2 | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 52 | 0.748 | 0.0437 | Yes |
| 2 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 58 | 0.726 | 0.0895 | Yes |
| 3 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 66 | 0.702 | 0.1337 | Yes |
| 4 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 95 | 0.621 | 0.1711 | Yes |
| 5 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 103 | 0.612 | 0.2095 | Yes |
| 6 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 143 | 0.545 | 0.2413 | Yes |
| 7 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 186 | 0.487 | 0.2691 | Yes |
| 8 | CARKL | CARKL Entrez, Source | carbohydrate kinase-like | 206 | 0.467 | 0.2974 | Yes |
| 9 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 247 | 0.438 | 0.3222 | Yes |
| 10 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.3486 | Yes |
| 11 | SLC2A5 | SLC2A5 Entrez, Source | solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | 276 | 0.417 | 0.3739 | Yes |
| 12 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 642 | 0.255 | 0.3626 | Yes |
| 13 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 666 | 0.249 | 0.3767 | Yes |
| 14 | SLC35A3 | SLC35A3 Entrez, Source | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 | 697 | 0.239 | 0.3896 | Yes |
| 15 | FUT8 | FUT8 Entrez, Source | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 762 | 0.225 | 0.3991 | Yes |
| 16 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.4055 | Yes |
| 17 | B4GALNT1 | B4GALNT1 Entrez, Source | beta-1,4-N-acetyl-galactosaminyl transferase 1 | 929 | 0.202 | 0.4131 | Yes |
| 18 | FUT1 | FUT1 Entrez, Source | fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) | 1004 | 0.192 | 0.4197 | Yes |
| 19 | GALK1 | GALK1 Entrez, Source | galactokinase 1 | 1032 | 0.189 | 0.4297 | Yes |
| 20 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 1047 | 0.187 | 0.4406 | Yes |
| 21 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 1183 | 0.172 | 0.4413 | Yes |
| 22 | NDST1 | NDST1 Entrez, Source | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 1209 | 0.170 | 0.4503 | Yes |
| 23 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 1243 | 0.168 | 0.4584 | Yes |
| 24 | ST8SIA4 | ST8SIA4 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | 1308 | 0.164 | 0.4640 | Yes |
| 25 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.4668 | Yes |
| 26 | ST8SIA2 | ST8SIA2 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 | 1450 | 0.153 | 0.4732 | Yes |
| 27 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 1462 | 0.152 | 0.4820 | Yes |
| 28 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 1469 | 0.151 | 0.4912 | Yes |
| 29 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1535 | 0.147 | 0.4956 | Yes |
| 30 | TFF1 | TFF1 Entrez, Source | trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) | 1578 | 0.144 | 0.5016 | Yes |
| 31 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 1617 | 0.142 | 0.5078 | Yes |
| 32 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1770 | 0.133 | 0.5048 | No |
| 33 | GALNT5 | GALNT5 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | 2282 | 0.111 | 0.4732 | No |
| 34 | TALDO1 | TALDO1 Entrez, Source | transaldolase 1 | 2443 | 0.105 | 0.4678 | No |
| 35 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 2527 | 0.102 | 0.4680 | No |
| 36 | ST3GAL5 | ST3GAL5 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | 2616 | 0.099 | 0.4676 | No |
| 37 | EPM2A | EPM2A Entrez, Source | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 2687 | 0.096 | 0.4685 | No |
| 38 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 2767 | 0.093 | 0.4684 | No |
| 39 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.4651 | No |
| 40 | INSR | INSR Entrez, Source | insulin receptor | 2897 | 0.088 | 0.4700 | No |
| 41 | NEU3 | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 2937 | 0.087 | 0.4726 | No |
| 42 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 2969 | 0.086 | 0.4757 | No |
| 43 | MPDU1 | MPDU1 Entrez, Source | mannose-P-dolichol utilization defect 1 | 3005 | 0.085 | 0.4784 | No |
| 44 | ARPP-19 | ARPP-19 Entrez, Source | - | 3013 | 0.084 | 0.4833 | No |
| 45 | SLC2A4 | SLC2A4 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 4 | 3091 | 0.082 | 0.4826 | No |
| 46 | GALNT7 | GALNT7 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 3152 | 0.080 | 0.4832 | No |
| 47 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 3168 | 0.079 | 0.4871 | No |
| 48 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 3278 | 0.077 | 0.4838 | No |
| 49 | ST6GAL1 | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 3300 | 0.076 | 0.4870 | No |
| 50 | ST6GALNAC6 | ST6GALNAC6 Entrez, Source | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 | 3421 | 0.073 | 0.4826 | No |
| 51 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 3429 | 0.073 | 0.4867 | No |
| 52 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 3545 | 0.070 | 0.4825 | No |
| 53 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.4599 | No |
| 54 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.4288 | No |
| 55 | XYLB | XYLB Entrez, Source | xylulokinase homolog (H. influenzae) | 4379 | 0.051 | 0.4302 | No |
| 56 | XYLT1 | XYLT1 Entrez, Source | xylosyltransferase I | 4421 | 0.050 | 0.4303 | No |
| 57 | ST8SIA5 | ST8SIA5 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 | 4440 | 0.050 | 0.4321 | No |
| 58 | B3GAT2 | B3GAT2 Entrez, Source | beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) | 4582 | 0.047 | 0.4244 | No |
| 59 | B4GALT7 | B4GALT7 Entrez, Source | xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) | 4905 | 0.040 | 0.4026 | No |
| 60 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 4971 | 0.038 | 0.4001 | No |
| 61 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5183 | 0.034 | 0.3864 | No |
| 62 | IPPK | IPPK Entrez, Source | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 5568 | 0.026 | 0.3590 | No |
| 63 | LECT1 | LECT1 Entrez, Source | leukocyte cell derived chemotaxin 1 | 5752 | 0.023 | 0.3467 | No |
| 64 | ADRB3 | ADRB3 Entrez, Source | adrenergic, beta-3-, receptor | 5857 | 0.021 | 0.3401 | No |
| 65 | FUT9 | FUT9 Entrez, Source | fucosyltransferase 9 (alpha (1,3) fucosyltransferase) | 5914 | 0.020 | 0.3372 | No |
| 66 | IL17F | IL17F Entrez, Source | interleukin 17F | 5953 | 0.019 | 0.3355 | No |
| 67 | CHST12 | CHST12 Entrez, Source | carbohydrate (chondroitin 4) sulfotransferase 12 | 6090 | 0.016 | 0.3263 | No |
| 68 | MGAT1 | MGAT1 Entrez, Source | mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | 6156 | 0.015 | 0.3224 | No |
| 69 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.3139 | No |
| 70 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 6397 | 0.010 | 0.3057 | No |
| 71 | PIGQ | PIGQ Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class Q | 6403 | 0.010 | 0.3059 | No |
| 72 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 6698 | 0.004 | 0.2840 | No |
| 73 | CHIA | CHIA Entrez, Source | chitinase, acidic | 6796 | 0.002 | 0.2768 | No |
| 74 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 6814 | 0.002 | 0.2756 | No |
| 75 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 7209 | -0.006 | 0.2463 | No |
| 76 | SLC5A2 | SLC5A2 Entrez, Source | solute carrier family 5 (sodium/glucose cotransporter), member 2 | 7237 | -0.007 | 0.2447 | No |
| 77 | FUT2 | FUT2 Entrez, Source | fucosyltransferase 2 (secretor status included) | 7831 | -0.019 | 0.2011 | No |
| 78 | ST6GAL2 | ST6GAL2 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | 7853 | -0.020 | 0.2008 | No |
| 79 | B3GAT1 | B3GAT1 Entrez, Source | beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) | 7886 | -0.021 | 0.1997 | No |
| 80 | NAGK | NAGK Entrez, Source | N-acetylglucosamine kinase | 7948 | -0.022 | 0.1965 | No |
| 81 | POMT1 | POMT1 Entrez, Source | protein-O-mannosyltransferase 1 | 8081 | -0.025 | 0.1881 | No |
| 82 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 8170 | -0.027 | 0.1832 | No |
| 83 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 8354 | -0.031 | 0.1714 | No |
| 84 | ST8SIA1 | ST8SIA1 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | 8578 | -0.036 | 0.1568 | No |
| 85 | LALBA | LALBA Entrez, Source | lactalbumin, alpha- | 8747 | -0.040 | 0.1467 | No |
| 86 | HPSE | HPSE Entrez, Source | heparanase | 8771 | -0.040 | 0.1475 | No |
| 87 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 8782 | -0.040 | 0.1493 | No |
| 88 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 8796 | -0.041 | 0.1509 | No |
| 89 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 9121 | -0.049 | 0.1296 | No |
| 90 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 9345 | -0.056 | 0.1163 | No |
| 91 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 9551 | -0.061 | 0.1047 | No |
| 92 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 9553 | -0.061 | 0.1085 | No |
| 93 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 9713 | -0.066 | 0.1006 | No |
| 94 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9745 | -0.066 | 0.1025 | No |
| 95 | GALT | GALT Entrez, Source | galactose-1-phosphate uridylyltransferase | 9817 | -0.069 | 0.1015 | No |
| 96 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 10142 | -0.079 | 0.0821 | No |
| 97 | FUT4 | FUT4 Entrez, Source | fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) | 10213 | -0.082 | 0.0820 | No |
| 98 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10469 | -0.091 | 0.0685 | No |
| 99 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 10743 | -0.100 | 0.0543 | No |
| 100 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 11033 | -0.113 | 0.0397 | No |
| 101 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 11815 | -0.158 | -0.0093 | No |
| 102 | ST8SIA3 | ST8SIA3 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | 11933 | -0.167 | -0.0075 | No |
| 103 | SLC2A8 | SLC2A8 Entrez, Source | solute carrier family 2, (facilitated glucose transporter) member 8 | 12054 | -0.178 | -0.0052 | No |
| 104 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 12474 | -0.224 | -0.0227 | No |
| 105 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12636 | -0.251 | -0.0188 | No |
| 106 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 12849 | -0.302 | -0.0156 | No |
| 107 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 13007 | -0.369 | -0.0040 | No |
| 108 | ST3GAL2 | ST3GAL2 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 13139 | -0.459 | 0.0153 | No |