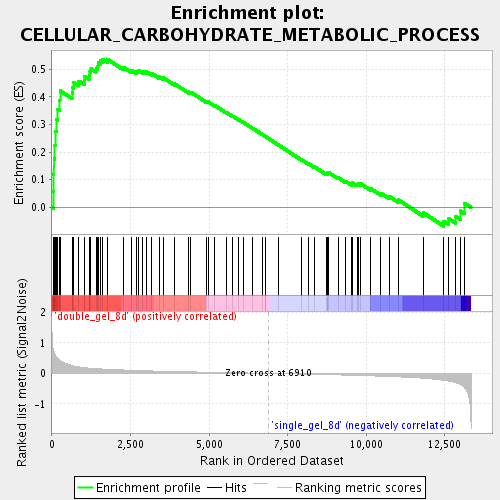
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CELLULAR_CARBOHYDRATE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5363253 |
| Normalized Enrichment Score (NES) | 1.8672383 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0072439415 |
| FWER p-Value | 0.605 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GYS2 | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 52 | 0.748 | 0.0583 | Yes |
| 2 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 58 | 0.726 | 0.1182 | Yes |
| 3 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 66 | 0.702 | 0.1761 | Yes |
| 4 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 95 | 0.621 | 0.2256 | Yes |
| 5 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 103 | 0.612 | 0.2759 | Yes |
| 6 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 143 | 0.545 | 0.3183 | Yes |
| 7 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 186 | 0.487 | 0.3556 | Yes |
| 8 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 247 | 0.438 | 0.3875 | Yes |
| 9 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.4222 | Yes |
| 10 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 642 | 0.255 | 0.4146 | Yes |
| 11 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 666 | 0.249 | 0.4336 | Yes |
| 12 | SLC35A3 | SLC35A3 Entrez, Source | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 | 697 | 0.239 | 0.4512 | Yes |
| 13 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.4568 | Yes |
| 14 | GALK1 | GALK1 Entrez, Source | galactokinase 1 | 1032 | 0.189 | 0.4595 | Yes |
| 15 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 1047 | 0.187 | 0.4740 | Yes |
| 16 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 1183 | 0.172 | 0.4782 | Yes |
| 17 | NDST1 | NDST1 Entrez, Source | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 1209 | 0.170 | 0.4904 | Yes |
| 18 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 1243 | 0.168 | 0.5018 | Yes |
| 19 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.5028 | Yes |
| 20 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 1462 | 0.152 | 0.5112 | Yes |
| 21 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 1469 | 0.151 | 0.5233 | Yes |
| 22 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1535 | 0.147 | 0.5306 | Yes |
| 23 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 1617 | 0.142 | 0.5363 | Yes |
| 24 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1770 | 0.133 | 0.5359 | No |
| 25 | GALNT5 | GALNT5 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | 2282 | 0.111 | 0.5066 | No |
| 26 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 2527 | 0.102 | 0.4967 | No |
| 27 | EPM2A | EPM2A Entrez, Source | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 2687 | 0.096 | 0.4927 | No |
| 28 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 2767 | 0.093 | 0.4945 | No |
| 29 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.4929 | No |
| 30 | ARPP-19 | ARPP-19 Entrez, Source | - | 3013 | 0.084 | 0.4905 | No |
| 31 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 3168 | 0.079 | 0.4855 | No |
| 32 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 3429 | 0.073 | 0.4720 | No |
| 33 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 3545 | 0.070 | 0.4691 | No |
| 34 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.4477 | No |
| 35 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.4178 | No |
| 36 | XYLT1 | XYLT1 Entrez, Source | xylosyltransferase I | 4421 | 0.050 | 0.4169 | No |
| 37 | B4GALT7 | B4GALT7 Entrez, Source | xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) | 4905 | 0.040 | 0.3838 | No |
| 38 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 4971 | 0.038 | 0.3821 | No |
| 39 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5183 | 0.034 | 0.3691 | No |
| 40 | IPPK | IPPK Entrez, Source | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 5568 | 0.026 | 0.3423 | No |
| 41 | LECT1 | LECT1 Entrez, Source | leukocyte cell derived chemotaxin 1 | 5752 | 0.023 | 0.3304 | No |
| 42 | IL17F | IL17F Entrez, Source | interleukin 17F | 5953 | 0.019 | 0.3170 | No |
| 43 | CHST12 | CHST12 Entrez, Source | carbohydrate (chondroitin 4) sulfotransferase 12 | 6090 | 0.016 | 0.3081 | No |
| 44 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 6397 | 0.010 | 0.2858 | No |
| 45 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 6698 | 0.004 | 0.2636 | No |
| 46 | CHIA | CHIA Entrez, Source | chitinase, acidic | 6796 | 0.002 | 0.2565 | No |
| 47 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 6814 | 0.002 | 0.2553 | No |
| 48 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 7209 | -0.006 | 0.2262 | No |
| 49 | NAGK | NAGK Entrez, Source | N-acetylglucosamine kinase | 7948 | -0.022 | 0.1724 | No |
| 50 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 8170 | -0.027 | 0.1580 | No |
| 51 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 8354 | -0.031 | 0.1468 | No |
| 52 | LALBA | LALBA Entrez, Source | lactalbumin, alpha- | 8747 | -0.040 | 0.1206 | No |
| 53 | HPSE | HPSE Entrez, Source | heparanase | 8771 | -0.040 | 0.1222 | No |
| 54 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 8782 | -0.040 | 0.1248 | No |
| 55 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 8796 | -0.041 | 0.1272 | No |
| 56 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 9121 | -0.049 | 0.1069 | No |
| 57 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 9345 | -0.056 | 0.0947 | No |
| 58 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 9551 | -0.061 | 0.0844 | No |
| 59 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 9553 | -0.061 | 0.0894 | No |
| 60 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 9713 | -0.066 | 0.0828 | No |
| 61 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9745 | -0.066 | 0.0860 | No |
| 62 | GALT | GALT Entrez, Source | galactose-1-phosphate uridylyltransferase | 9817 | -0.069 | 0.0864 | No |
| 63 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 10142 | -0.079 | 0.0686 | No |
| 64 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10469 | -0.091 | 0.0515 | No |
| 65 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 10743 | -0.100 | 0.0393 | No |
| 66 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 11033 | -0.113 | 0.0270 | No |
| 67 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 11815 | -0.158 | -0.0188 | No |
| 68 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 12474 | -0.224 | -0.0498 | No |
| 69 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12636 | -0.251 | -0.0410 | No |
| 70 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 12849 | -0.302 | -0.0319 | No |
| 71 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 13007 | -0.369 | -0.0130 | No |
| 72 | ST3GAL2 | ST3GAL2 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 13139 | -0.459 | 0.0153 | No |

