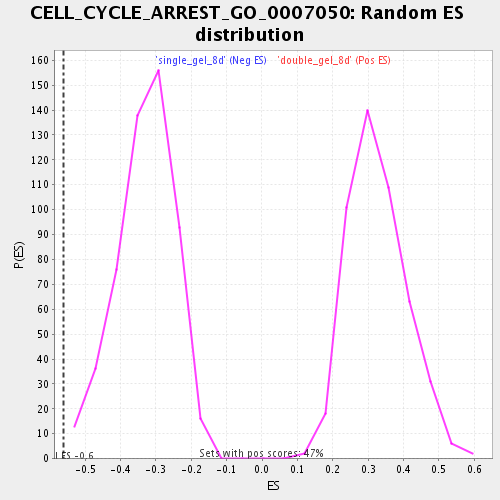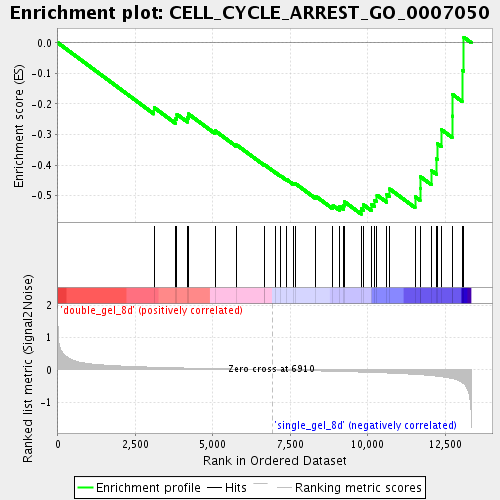
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | CELL_CYCLE_ARREST_GO_0007050 |
| Enrichment Score (ES) | -0.5607345 |
| Normalized Enrichment Score (NES) | -1.7051384 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.075398155 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TBRG4 | TBRG4 Entrez, Source | transforming growth factor beta regulator 4 | 3101 | 0.081 | -0.2117 | No |
| 2 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 3805 | 0.064 | -0.2479 | No |
| 3 | AIF1 | AIF1 Entrez, Source | allograft inflammatory factor 1 | 3839 | 0.063 | -0.2338 | No |
| 4 | INHA | INHA Entrez, Source | inhibin, alpha | 4183 | 0.055 | -0.2451 | No |
| 5 | PCAF | PCAF Entrez, Source | p300/CBP-associated factor | 4209 | 0.055 | -0.2326 | No |
| 6 | GAS7 | GAS7 Entrez, Source | growth arrest-specific 7 | 5073 | 0.036 | -0.2879 | No |
| 7 | BTG4 | BTG4 Entrez, Source | B-cell translocation gene 4 | 5769 | 0.022 | -0.3343 | No |
| 8 | UHMK1 | UHMK1 Entrez, Source | U2AF homology motif (UHM) kinase 1 | 6651 | 0.005 | -0.3991 | No |
| 9 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 6668 | 0.005 | -0.3991 | No |
| 10 | PPM1G | PPM1G Entrez, Source | protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform | 7035 | -0.003 | -0.4259 | No |
| 11 | MFN2 | MFN2 Entrez, Source | mitofusin 2 | 7196 | -0.006 | -0.4363 | No |
| 12 | INHBA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 7375 | -0.010 | -0.4471 | No |
| 13 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 7589 | -0.014 | -0.4594 | No |
| 14 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 7666 | -0.016 | -0.4609 | No |
| 15 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 8322 | -0.030 | -0.5022 | No |
| 16 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 8877 | -0.043 | -0.5326 | No |
| 17 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9083 | -0.048 | -0.5354 | No |
| 18 | CUL5 | CUL5 Entrez, Source | cullin 5 | 9220 | -0.052 | -0.5320 | No |
| 19 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 9252 | -0.053 | -0.5205 | No |
| 20 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9788 | -0.068 | -0.5431 | Yes |
| 21 | PPP1R9B | PPP1R9B Entrez, Source | protein phosphatase 1, regulatory subunit 9B, spinophilin | 9861 | -0.070 | -0.5301 | Yes |
| 22 | NBN | NBN Entrez, Source | nibrin | 10122 | -0.079 | -0.5290 | Yes |
| 23 | APBB1 | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 10220 | -0.082 | -0.5149 | Yes |
| 24 | APC | APC Entrez, Source | adenomatosis polyposis coli | 10296 | -0.085 | -0.4984 | Yes |
| 25 | RASSF1 | RASSF1 Entrez, Source | Ras association (RalGDS/AF-6) domain family 1 | 10613 | -0.096 | -0.4971 | Yes |
| 26 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 10707 | -0.099 | -0.4783 | Yes |
| 27 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11527 | -0.139 | -0.5035 | Yes |
| 28 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11690 | -0.149 | -0.4767 | Yes |
| 29 | TBRG1 | TBRG1 Entrez, Source | transforming growth factor beta regulator 1 | 11701 | -0.150 | -0.4383 | Yes |
| 30 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 12056 | -0.178 | -0.4184 | Yes |
| 31 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 12219 | -0.194 | -0.3799 | Yes |
| 32 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 12251 | -0.199 | -0.3302 | Yes |
| 33 | ING4 | ING4 Entrez, Source | inhibitor of growth family, member 4 | 12380 | -0.212 | -0.2843 | Yes |
| 34 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 12725 | -0.270 | -0.2395 | Yes |
| 35 | NOTCH2 | NOTCH2 Entrez, Source | Notch homolog 2 (Drosophila) | 12734 | -0.272 | -0.1688 | Yes |
| 36 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 13047 | -0.395 | -0.0890 | Yes |
| 37 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 13099 | -0.425 | 0.0183 | Yes |