Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
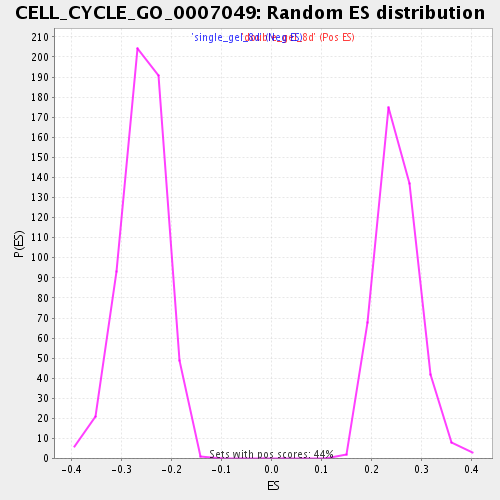
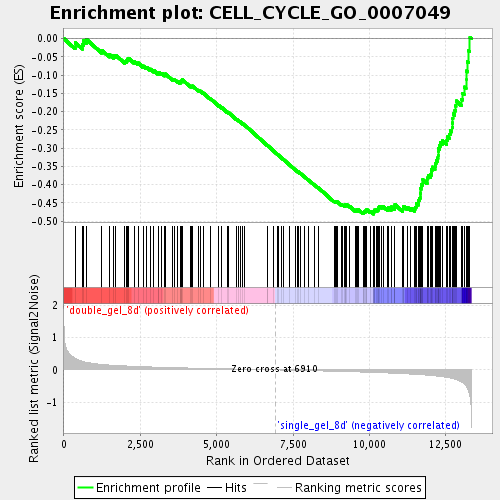
| GeneSet | CELL_CYCLE_GO_0007049 |
| Enrichment Score (ES) | -0.48063856 |
| Normalized Enrichment Score (NES) | -1.8628359 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.029655136 |
| FWER p-Value | 0.655 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BMP2 | BMP2 Entrez, Source | bone morphogenetic protein 2 | 370 | 0.355 | -0.0108 | No |
| 2 | MDM4 | MDM4 Entrez, Source | Mdm4, transformed 3T3 cell double minute 4, p53 binding protein (mouse) | 605 | 0.265 | -0.0156 | No |
| 3 | BMP7 | BMP7 Entrez, Source | bone morphogenetic protein 7 (osteogenic protein 1) | 635 | 0.257 | -0.0053 | No |
| 4 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 740 | 0.230 | -0.0019 | No |
| 5 | PTPRC | PTPRC Entrez, Source | protein tyrosine phosphatase, receptor type, C | 1238 | 0.168 | -0.0315 | No |
| 6 | EGFL6 | EGFL6 Entrez, Source | EGF-like-domain, multiple 6 | 1486 | 0.151 | -0.0429 | No |
| 7 | CDK5R1 | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 1630 | 0.141 | -0.0468 | No |
| 8 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1695 | 0.137 | -0.0450 | No |
| 9 | SPDYA | SPDYA Entrez, Source | speedy homolog A (Drosophila) | 1994 | 0.122 | -0.0616 | No |
| 10 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 2055 | 0.119 | -0.0604 | No |
| 11 | CDK6 | CDK6 Entrez, Source | cyclin-dependent kinase 6 | 2067 | 0.119 | -0.0554 | No |
| 12 | BMP4 | BMP4 Entrez, Source | bone morphogenetic protein 4 | 2116 | 0.117 | -0.0534 | No |
| 13 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 2307 | 0.110 | -0.0624 | No |
| 14 | ASNS | ASNS Entrez, Source | asparagine synthetase | 2424 | 0.106 | -0.0661 | No |
| 15 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 2601 | 0.099 | -0.0746 | No |
| 16 | RUNX3 | RUNX3 Entrez, Source | runt-related transcription factor 3 | 2707 | 0.095 | -0.0779 | No |
| 17 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 2832 | 0.090 | -0.0829 | No |
| 18 | PPP5C | PPP5C Entrez, Source | protein phosphatase 5, catalytic subunit | 2945 | 0.087 | -0.0872 | No |
| 19 | TBRG4 | TBRG4 Entrez, Source | transforming growth factor beta regulator 4 | 3101 | 0.081 | -0.0950 | No |
| 20 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3109 | 0.081 | -0.0915 | No |
| 21 | SUGT1 | SUGT1 Entrez, Source | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | 3198 | 0.079 | -0.0944 | No |
| 22 | USH1C | USH1C Entrez, Source | Usher syndrome 1C (autosomal recessive, severe) | 3303 | 0.076 | -0.0986 | No |
| 23 | SYCP1 | SYCP1 Entrez, Source | synaptonemal complex protein 1 | 3323 | 0.075 | -0.0963 | No |
| 24 | CDK5RAP1 | CDK5RAP1 Entrez, Source | CDK5 regulatory subunit associated protein 1 | 3568 | 0.069 | -0.1114 | No |
| 25 | EREG | EREG Entrez, Source | epiregulin | 3625 | 0.068 | -0.1124 | No |
| 26 | KPNA2 | KPNA2 Entrez, Source | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 3717 | 0.066 | -0.1161 | No |
| 27 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 3805 | 0.064 | -0.1195 | No |
| 28 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 3808 | 0.064 | -0.1166 | No |
| 29 | AIF1 | AIF1 Entrez, Source | allograft inflammatory factor 1 | 3839 | 0.063 | -0.1158 | No |
| 30 | CENTD2 | CENTD2 Entrez, Source | centaurin, delta 2 | 3860 | 0.063 | -0.1142 | No |
| 31 | RHOB | RHOB Entrez, Source | ras homolog gene family, member B | 3872 | 0.063 | -0.1120 | No |
| 32 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 4143 | 0.056 | -0.1298 | No |
| 33 | INHA | INHA Entrez, Source | inhibin, alpha | 4183 | 0.055 | -0.1300 | No |
| 34 | PCAF | PCAF Entrez, Source | p300/CBP-associated factor | 4209 | 0.055 | -0.1292 | No |
| 35 | TTN | TTN Entrez, Source | titin | 4401 | 0.050 | -0.1413 | No |
| 36 | CDK5RAP3 | CDK5RAP3 Entrez, Source | CDK5 regulatory subunit associated protein 3 | 4459 | 0.049 | -0.1432 | No |
| 37 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 4562 | 0.047 | -0.1486 | No |
| 38 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4801 | 0.042 | -0.1646 | No |
| 39 | GAS7 | GAS7 Entrez, Source | growth arrest-specific 7 | 5073 | 0.036 | -0.1834 | No |
| 40 | STAG3 | STAG3 Entrez, Source | stromal antigen 3 | 5167 | 0.035 | -0.1888 | No |
| 41 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 5345 | 0.031 | -0.2007 | No |
| 42 | NEK6 | NEK6 Entrez, Source | NIMA (never in mitosis gene a)-related kinase 6 | 5393 | 0.030 | -0.2028 | No |
| 43 | GFI1 | GFI1 Entrez, Source | growth factor independent 1 | 5640 | 0.025 | -0.2202 | No |
| 44 | DUSP13 | DUSP13 Entrez, Source | dual specificity phosphatase 13 | 5702 | 0.024 | -0.2237 | No |
| 45 | BTG4 | BTG4 Entrez, Source | B-cell translocation gene 4 | 5769 | 0.022 | -0.2276 | No |
| 46 | ZBTB17 | ZBTB17 Entrez, Source | zinc finger and BTB domain containing 17 | 5859 | 0.021 | -0.2333 | No |
| 47 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 5925 | 0.020 | -0.2373 | No |
| 48 | UHMK1 | UHMK1 Entrez, Source | U2AF homology motif (UHM) kinase 1 | 6651 | 0.005 | -0.2920 | No |
| 49 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 6668 | 0.005 | -0.2930 | No |
| 50 | CETN3 | CETN3 Entrez, Source | centrin, EF-hand protein, 3 (CDC31 homolog, yeast) | 6856 | 0.001 | -0.3071 | No |
| 51 | SNF1LK | SNF1LK Entrez, Source | SNF1-like kinase | 6978 | -0.002 | -0.3162 | No |
| 52 | PPM1G | PPM1G Entrez, Source | protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform | 7035 | -0.003 | -0.3203 | No |
| 53 | SERTAD1 | SERTAD1 Entrez, Source | SERTA domain containing 1 | 7125 | -0.005 | -0.3269 | No |
| 54 | MFN2 | MFN2 Entrez, Source | mitofusin 2 | 7196 | -0.006 | -0.3319 | No |
| 55 | INHBA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 7375 | -0.010 | -0.3449 | No |
| 56 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 7589 | -0.014 | -0.3604 | No |
| 57 | GTPBP4 | GTPBP4 Entrez, Source | GTP binding protein 4 | 7656 | -0.016 | -0.3646 | No |
| 58 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 7666 | -0.016 | -0.3645 | No |
| 59 | KATNA1 | KATNA1 Entrez, Source | katanin p60 (ATPase-containing) subunit A 1 | 7746 | -0.018 | -0.3697 | No |
| 60 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 7859 | -0.020 | -0.3772 | No |
| 61 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 8008 | -0.024 | -0.3873 | No |
| 62 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 8204 | -0.028 | -0.4007 | No |
| 63 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 8322 | -0.030 | -0.4081 | No |
| 64 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 8842 | -0.042 | -0.4454 | No |
| 65 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 8877 | -0.043 | -0.4459 | No |
| 66 | PPP6C | PPP6C Entrez, Source | protein phosphatase 6, catalytic subunit | 8915 | -0.044 | -0.4466 | No |
| 67 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 8949 | -0.045 | -0.4469 | No |
| 68 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9083 | -0.048 | -0.4546 | No |
| 69 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 9128 | -0.050 | -0.4555 | No |
| 70 | NPM2 | NPM2 Entrez, Source | nucleophosmin/nucleoplasmin, 2 | 9187 | -0.051 | -0.4575 | No |
| 71 | GPS1 | GPS1 Entrez, Source | G protein pathway suppressor 1 | 9208 | -0.051 | -0.4565 | No |
| 72 | CUL5 | CUL5 Entrez, Source | cullin 5 | 9220 | -0.052 | -0.4548 | No |
| 73 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 9252 | -0.053 | -0.4546 | No |
| 74 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 9345 | -0.056 | -0.4588 | No |
| 75 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 9534 | -0.061 | -0.4701 | No |
| 76 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 9560 | -0.061 | -0.4690 | No |
| 77 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 9617 | -0.063 | -0.4702 | No |
| 78 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 9627 | -0.063 | -0.4678 | No |
| 79 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9788 | -0.068 | -0.4766 | No |
| 80 | ANAPC4 | ANAPC4 Entrez, Source | anaphase promoting complex subunit 4 | 9814 | -0.069 | -0.4752 | No |
| 81 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 9827 | -0.069 | -0.4727 | No |
| 82 | PPP1R9B | PPP1R9B Entrez, Source | protein phosphatase 1, regulatory subunit 9B, spinophilin | 9861 | -0.070 | -0.4718 | No |
| 83 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 9894 | -0.071 | -0.4707 | No |
| 84 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9912 | -0.072 | -0.4685 | No |
| 85 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 10026 | -0.075 | -0.4734 | No |
| 86 | NBN | NBN Entrez, Source | nibrin | 10122 | -0.079 | -0.4768 | Yes |
| 87 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 10127 | -0.079 | -0.4732 | Yes |
| 88 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 10151 | -0.080 | -0.4711 | Yes |
| 89 | UBB | UBB Entrez, Source | ubiquitin B | 10179 | -0.081 | -0.4692 | Yes |
| 90 | APBB1 | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 10220 | -0.082 | -0.4683 | Yes |
| 91 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 10276 | -0.084 | -0.4683 | Yes |
| 92 | APC | APC Entrez, Source | adenomatosis polyposis coli | 10296 | -0.085 | -0.4657 | Yes |
| 93 | DCTN2 | DCTN2 Entrez, Source | dynactin 2 (p50) | 10303 | -0.085 | -0.4620 | Yes |
| 94 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 10312 | -0.085 | -0.4585 | Yes |
| 95 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 10391 | -0.088 | -0.4601 | Yes |
| 96 | SC65 | SC65 Entrez, Source | - | 10443 | -0.090 | -0.4596 | Yes |
| 97 | CETN1 | CETN1 Entrez, Source | centrin, EF-hand protein, 1 | 10599 | -0.095 | -0.4667 | Yes |
| 98 | RASSF1 | RASSF1 Entrez, Source | Ras association (RalGDS/AF-6) domain family 1 | 10613 | -0.096 | -0.4630 | Yes |
| 99 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 10707 | -0.099 | -0.4653 | Yes |
| 100 | CHFR | CHFR Entrez, Source | checkpoint with forkhead and ring finger domains | 10712 | -0.099 | -0.4608 | Yes |
| 101 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 10819 | -0.104 | -0.4637 | Yes |
| 102 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 10825 | -0.104 | -0.4590 | Yes |
| 103 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 10832 | -0.104 | -0.4544 | Yes |
| 104 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 11087 | -0.116 | -0.4680 | Yes |
| 105 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11099 | -0.117 | -0.4632 | Yes |
| 106 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 11117 | -0.118 | -0.4587 | Yes |
| 107 | AKAP8 | AKAP8 Entrez, Source | A kinase (PRKA) anchor protein 8 | 11248 | -0.124 | -0.4626 | Yes |
| 108 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 11352 | -0.129 | -0.4641 | Yes |
| 109 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 11467 | -0.135 | -0.4661 | Yes |
| 110 | CDK10 | CDK10 Entrez, Source | cyclin-dependent kinase (CDC2-like) 10 | 11493 | -0.137 | -0.4614 | Yes |
| 111 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11527 | -0.139 | -0.4571 | Yes |
| 112 | AHR | AHR Entrez, Source | aryl hydrocarbon receptor | 11529 | -0.139 | -0.4504 | Yes |
| 113 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 11603 | -0.144 | -0.4489 | Yes |
| 114 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 11608 | -0.144 | -0.4422 | Yes |
| 115 | CIT | CIT Entrez, Source | citron (rho-interacting, serine/threonine kinase 21) | 11639 | -0.146 | -0.4374 | Yes |
| 116 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11658 | -0.148 | -0.4315 | Yes |
| 117 | BTG3 | BTG3 Entrez, Source | BTG family, member 3 | 11661 | -0.148 | -0.4245 | Yes |
| 118 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11672 | -0.148 | -0.4180 | Yes |
| 119 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 11673 | -0.148 | -0.4108 | Yes |
| 120 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11690 | -0.149 | -0.4048 | Yes |
| 121 | TBRG1 | TBRG1 Entrez, Source | transforming growth factor beta regulator 1 | 11701 | -0.150 | -0.3982 | Yes |
| 122 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 11744 | -0.152 | -0.3940 | Yes |
| 123 | CD28 | CD28 Entrez, Source | CD28 molecule | 11745 | -0.152 | -0.3866 | Yes |
| 124 | DDB1 | DDB1 Entrez, Source | damage-specific DNA binding protein 1, 127kDa | 11887 | -0.163 | -0.3893 | Yes |
| 125 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 11889 | -0.164 | -0.3814 | Yes |
| 126 | SMC4 | SMC4 Entrez, Source | structural maintenance of chromosomes 4 | 11916 | -0.167 | -0.3753 | Yes |
| 127 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 11998 | -0.173 | -0.3730 | Yes |
| 128 | SMC3 | SMC3 Entrez, Source | structural maintenance of chromosomes 3 | 12021 | -0.175 | -0.3662 | Yes |
| 129 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 12031 | -0.176 | -0.3583 | Yes |
| 130 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 12056 | -0.178 | -0.3514 | Yes |
| 131 | HSPA2 | HSPA2 Entrez, Source | heat shock 70kDa protein 2 | 12146 | -0.186 | -0.3491 | Yes |
| 132 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 12163 | -0.188 | -0.3412 | Yes |
| 133 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 12182 | -0.190 | -0.3333 | Yes |
| 134 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 12219 | -0.194 | -0.3266 | Yes |
| 135 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 12243 | -0.198 | -0.3187 | Yes |
| 136 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 12251 | -0.199 | -0.3095 | Yes |
| 137 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 12258 | -0.199 | -0.3003 | Yes |
| 138 | CDC37 | CDC37 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae) | 12294 | -0.204 | -0.2930 | Yes |
| 139 | SH3BP4 | SH3BP4 Entrez, Source | SH3-domain binding protein 4 | 12316 | -0.206 | -0.2845 | Yes |
| 140 | ING4 | ING4 Entrez, Source | inhibitor of growth family, member 4 | 12380 | -0.212 | -0.2790 | Yes |
| 141 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 12531 | -0.233 | -0.2790 | Yes |
| 142 | AURKA | AURKA Entrez, Source | aurora kinase A | 12548 | -0.235 | -0.2687 | Yes |
| 143 | CKAP5 | CKAP5 Entrez, Source | cytoskeleton associated protein 5 | 12613 | -0.247 | -0.2615 | Yes |
| 144 | TIMELESS | TIMELESS Entrez, Source | timeless homolog (Drosophila) | 12644 | -0.253 | -0.2515 | Yes |
| 145 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12700 | -0.264 | -0.2428 | Yes |
| 146 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 12713 | -0.267 | -0.2307 | Yes |
| 147 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 12725 | -0.270 | -0.2183 | Yes |
| 148 | NOTCH2 | NOTCH2 Entrez, Source | Notch homolog 2 (Drosophila) | 12734 | -0.272 | -0.2057 | Yes |
| 149 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 12794 | -0.284 | -0.1963 | Yes |
| 150 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12821 | -0.294 | -0.1839 | Yes |
| 151 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12832 | -0.296 | -0.1703 | Yes |
| 152 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13016 | -0.374 | -0.1659 | Yes |
| 153 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 13047 | -0.395 | -0.1489 | Yes |
| 154 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 13099 | -0.425 | -0.1321 | Yes |
| 155 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13162 | -0.500 | -0.1125 | Yes |
| 156 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13167 | -0.505 | -0.0882 | Yes |
| 157 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 13208 | -0.576 | -0.0631 | Yes |
| 158 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13247 | -0.677 | -0.0330 | Yes |
| 159 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13288 | -0.823 | 0.0041 | Yes |