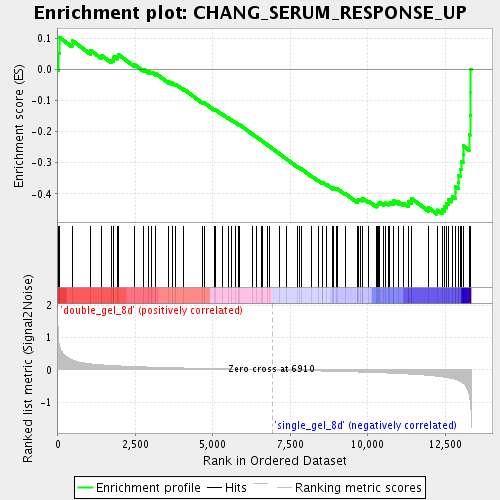
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | CHANG_SERUM_RESPONSE_UP |
| Enrichment Score (ES) | -0.4651093 |
| Normalized Enrichment Score (NES) | -1.6527703 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.096198514 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAP3K8 | MAP3K8 Entrez, Source | mitogen-activated protein kinase kinase kinase 8 | 34 | 0.817 | 0.0522 | No |
| 2 | PLG | PLG Entrez, Source | plasminogen | 44 | 0.773 | 0.1033 | No |
| 3 | DHFR | DHFR Entrez, Source | dihydrofolate reductase | 457 | 0.317 | 0.0935 | No |
| 4 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 1050 | 0.187 | 0.0614 | No |
| 5 | EIF4EBP1 | EIF4EBP1 Entrez, Source | eukaryotic translation initiation factor 4E binding protein 1 | 1395 | 0.157 | 0.0460 | No |
| 6 | MKKS | MKKS Entrez, Source | McKusick-Kaufman syndrome | 1728 | 0.135 | 0.0300 | No |
| 7 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 1785 | 0.132 | 0.0346 | No |
| 8 | SRM | SRM Entrez, Source | spermidine synthase | 1809 | 0.131 | 0.0416 | No |
| 9 | COX17 | COX17 Entrez, Source | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) | 1921 | 0.125 | 0.0417 | No |
| 10 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 1940 | 0.124 | 0.0486 | No |
| 11 | LYAR | LYAR Entrez, Source | - | 2462 | 0.104 | 0.0163 | No |
| 12 | SLC25A5 | SLC25A5 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 2756 | 0.094 | 0.0005 | No |
| 13 | SLC16A1 | SLC16A1 Entrez, Source | solute carrier family 16, member 1 (monocarboxylic acid transporter 1) | 2935 | 0.087 | -0.0071 | No |
| 14 | MRPL37 | MRPL37 Entrez, Source | mitochondrial ribosomal protein L37 | 3031 | 0.084 | -0.0087 | No |
| 15 | TPI1 | TPI1 Entrez, Source | triosephosphate isomerase 1 | 3163 | 0.080 | -0.0132 | No |
| 16 | FLNC | FLNC Entrez, Source | filamin C, gamma (actin binding protein 280) | 3577 | 0.069 | -0.0397 | No |
| 17 | SSR3 | SSR3 Entrez, Source | signal sequence receptor, gamma (translocon-associated protein gamma) | 3688 | 0.067 | -0.0436 | No |
| 18 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 3808 | 0.064 | -0.0483 | No |
| 19 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 4054 | 0.058 | -0.0628 | No |
| 20 | MT3 | MT3 Entrez, Source | metallothionein 3 (growth inhibitory factor (neurotrophic)) | 4680 | 0.044 | -0.1070 | No |
| 21 | NUDT1 | NUDT1 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 1 | 4723 | 0.043 | -0.1073 | No |
| 22 | CENPJ | CENPJ Entrez, Source | centromere protein J | 5047 | 0.037 | -0.1292 | No |
| 23 | SNRPA | SNRPA Entrez, Source | small nuclear ribonucleoprotein polypeptide A | 5088 | 0.036 | -0.1298 | No |
| 24 | TFPI2 | TFPI2 Entrez, Source | tissue factor pathway inhibitor 2 | 5317 | 0.032 | -0.1449 | No |
| 25 | PCSK7 | PCSK7 Entrez, Source | proprotein convertase subtilisin/kexin type 7 | 5492 | 0.028 | -0.1561 | No |
| 26 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 5597 | 0.026 | -0.1622 | No |
| 27 | TCEB1 | TCEB1 Entrez, Source | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | 5725 | 0.023 | -0.1703 | No |
| 28 | RPN1 | RPN1 Entrez, Source | ribophorin I | 5839 | 0.021 | -0.1774 | No |
| 29 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 5852 | 0.021 | -0.1769 | No |
| 30 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 6264 | 0.013 | -0.2070 | No |
| 31 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 6421 | 0.009 | -0.2182 | No |
| 32 | IMP4 | IMP4 Entrez, Source | IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) | 6557 | 0.007 | -0.2279 | No |
| 33 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 6598 | 0.006 | -0.2305 | No |
| 34 | PSMA7 | PSMA7 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 7 | 6760 | 0.003 | -0.2425 | No |
| 35 | WSB2 | WSB2 Entrez, Source | WD repeat and SOCS box-containing 2 | 6837 | 0.002 | -0.2481 | No |
| 36 | UMPS | UMPS Entrez, Source | uridine monophosphate synthetase (orotate phosphoribosyl transferase and orotidine-5'-decarboxylase) | 7149 | -0.005 | -0.2712 | No |
| 37 | GPLD1 | GPLD1 Entrez, Source | glycosylphosphatidylinositol specific phospholipase D1 | 7374 | -0.010 | -0.2875 | No |
| 38 | EMP2 | EMP2 Entrez, Source | epithelial membrane protein 2 | 7732 | -0.017 | -0.3133 | No |
| 39 | NLN | NLN Entrez, Source | neurolysin (metallopeptidase M3 family) | 7790 | -0.018 | -0.3163 | No |
| 40 | TNFRSF12A | TNFRSF12A Entrez, Source | tumor necrosis factor receptor superfamily, member 12A | 7849 | -0.020 | -0.3194 | No |
| 41 | CDCA4 | CDCA4 Entrez, Source | cell division cycle associated 4 | 8197 | -0.027 | -0.3438 | No |
| 42 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 8399 | -0.032 | -0.3567 | No |
| 43 | PDAP1 | PDAP1 Entrez, Source | PDGFA associated protein 1 | 8528 | -0.035 | -0.3640 | No |
| 44 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 8540 | -0.035 | -0.3625 | No |
| 45 | PLAUR | PLAUR Entrez, Source | plasminogen activator, urokinase receptor | 8677 | -0.038 | -0.3702 | No |
| 46 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 8877 | -0.043 | -0.3823 | No |
| 47 | EBNA1BP2 | EBNA1BP2 Entrez, Source | EBNA1 binding protein 2 | 8885 | -0.043 | -0.3799 | No |
| 48 | NUTF2 | NUTF2 Entrez, Source | nuclear transport factor 2 | 8981 | -0.045 | -0.3841 | No |
| 49 | PSMD12 | PSMD12 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 9015 | -0.046 | -0.3835 | No |
| 50 | PSMD2 | PSMD2 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 9272 | -0.054 | -0.3992 | No |
| 51 | RNF138 | RNF138 Entrez, Source | ring finger protein 138 | 9660 | -0.064 | -0.4241 | No |
| 52 | GNG11 | GNG11 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 11 | 9666 | -0.064 | -0.4202 | No |
| 53 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 9695 | -0.065 | -0.4179 | No |
| 54 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 9762 | -0.067 | -0.4184 | No |
| 55 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 9827 | -0.069 | -0.4186 | No |
| 56 | HAS2 | HAS2 Entrez, Source | hyaluronan synthase 2 | 9840 | -0.070 | -0.4148 | No |
| 57 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 10026 | -0.075 | -0.4238 | No |
| 58 | RUVBL1 | RUVBL1 Entrez, Source | RuvB-like 1 (E. coli) | 10274 | -0.084 | -0.4368 | No |
| 59 | TXNL2 | TXNL2 Entrez, Source | thioredoxin-like 2 | 10307 | -0.085 | -0.4335 | No |
| 60 | PAICS | PAICS Entrez, Source | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 10338 | -0.086 | -0.4300 | No |
| 61 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 10369 | -0.087 | -0.4264 | No |
| 62 | NUP107 | NUP107 Entrez, Source | nucleoporin 107kDa | 10509 | -0.092 | -0.4308 | No |
| 63 | RNASEH2A | RNASEH2A Entrez, Source | ribonuclease H2, subunit A | 10571 | -0.094 | -0.4291 | No |
| 64 | ADAMTS1 | ADAMTS1 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 10664 | -0.097 | -0.4295 | No |
| 65 | RNF41 | RNF41 Entrez, Source | ring finger protein 41 | 10715 | -0.099 | -0.4266 | No |
| 66 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 10832 | -0.104 | -0.4284 | No |
| 67 | CCT5 | CCT5 Entrez, Source | chaperonin containing TCP1, subunit 5 (epsilon) | 10843 | -0.105 | -0.4221 | No |
| 68 | PSMC3 | PSMC3 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 10989 | -0.111 | -0.4256 | No |
| 69 | PFN1 | PFN1 Entrez, Source | profilin 1 | 11162 | -0.120 | -0.4306 | No |
| 70 | SNRPB | SNRPB Entrez, Source | small nuclear ribonucleoprotein polypeptides B and B1 | 11304 | -0.127 | -0.4328 | No |
| 71 | HNRPR | HNRPR Entrez, Source | heterogeneous nuclear ribonucleoprotein R | 11314 | -0.127 | -0.4249 | No |
| 72 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 11400 | -0.131 | -0.4225 | No |
| 73 | DCK | DCK Entrez, Source | deoxycytidine kinase | 11406 | -0.131 | -0.4141 | No |
| 74 | NUPL1 | NUPL1 Entrez, Source | nucleoporin like 1 | 11959 | -0.169 | -0.4444 | No |
| 75 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 12234 | -0.197 | -0.4519 | Yes |
| 76 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 12405 | -0.215 | -0.4503 | Yes |
| 77 | DCLRE1B | DCLRE1B Entrez, Source | DNA cross-link repair 1B (PSO2 homolog, S. cerevisiae) | 12486 | -0.225 | -0.4412 | Yes |
| 78 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 12545 | -0.235 | -0.4299 | Yes |
| 79 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12611 | -0.247 | -0.4182 | Yes |
| 80 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 12719 | -0.269 | -0.4082 | Yes |
| 81 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12821 | -0.294 | -0.3962 | Yes |
| 82 | GGH | GGH Entrez, Source | gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 12824 | -0.294 | -0.3766 | Yes |
| 83 | MET | MET Entrez, Source | met proto-oncogene (hepatocyte growth factor receptor) | 12926 | -0.331 | -0.3621 | Yes |
| 84 | CKLF | CKLF Entrez, Source | chemokine-like factor | 12938 | -0.335 | -0.3404 | Yes |
| 85 | TPM1 | TPM1 Entrez, Source | tropomyosin 1 (alpha) | 12998 | -0.364 | -0.3205 | Yes |
| 86 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13028 | -0.381 | -0.2972 | Yes |
| 87 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 13077 | -0.411 | -0.2732 | Yes |
| 88 | SMS | SMS Entrez, Source | spermine synthase | 13083 | -0.414 | -0.2458 | Yes |
| 89 | VIL2 | VIL2 Entrez, Source | villin 2 (ezrin) | 13271 | -0.763 | -0.2088 | Yes |
| 90 | MSN | MSN Entrez, Source | moesin | 13302 | -0.941 | -0.1479 | Yes |
| 91 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 13324 | -1.119 | -0.0745 | Yes |
| 92 | TAGLN | TAGLN Entrez, Source | transgelin | 13326 | -1.131 | 0.0012 | Yes |