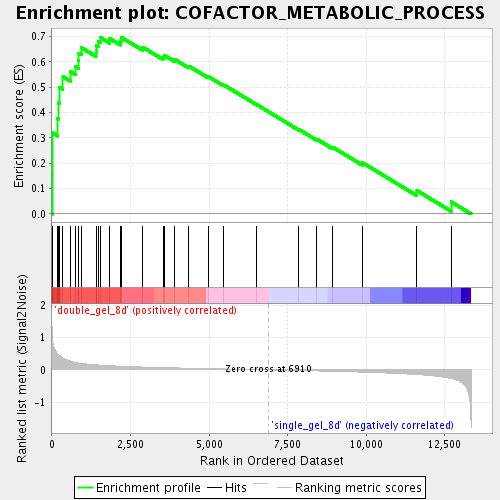
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | COFACTOR_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.6967209 |
| Normalized Enrichment Score (NES) | 2.0520728 |
| Nominal p-value | 0.0 |
| FDR q-value | 9.573071E-4 |
| FWER p-Value | 0.069 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FTCD | FTCD Entrez, Source | formiminotransferase cyclodeaminase | 9 | 1.171 | 0.1633 | Yes |
| 2 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 10 | 1.121 | 0.3203 | Yes |
| 3 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 186 | 0.487 | 0.3753 | Yes |
| 4 | ACOT12 | ACOT12 Entrez, Source | acyl-CoA thioesterase 12 | 212 | 0.461 | 0.4380 | Yes |
| 5 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 231 | 0.450 | 0.4997 | Yes |
| 6 | MOCS2 | MOCS2 Entrez, Source | molybdenum cofactor synthesis 2 | 352 | 0.361 | 0.5412 | Yes |
| 7 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 584 | 0.271 | 0.5619 | Yes |
| 8 | ALDH1L1 | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 750 | 0.228 | 0.5813 | Yes |
| 9 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 839 | 0.215 | 0.6048 | Yes |
| 10 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.6333 | Yes |
| 11 | COQ3 | COQ3 Entrez, Source | coenzyme Q3 homolog, methyltransferase (S. cerevisiae) | 932 | 0.202 | 0.6561 | Yes |
| 12 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.6426 | Yes |
| 13 | ALAD | ALAD Entrez, Source | aminolevulinate, delta-, dehydratase | 1424 | 0.155 | 0.6630 | Yes |
| 14 | COQ7 | COQ7 Entrez, Source | coenzyme Q7 homolog, ubiquinone (yeast) | 1476 | 0.151 | 0.6803 | Yes |
| 15 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1535 | 0.147 | 0.6965 | Yes |
| 16 | UROS | UROS Entrez, Source | uroporphyrinogen III synthase (congenital erythropoietic porphyria) | 1842 | 0.129 | 0.6916 | Yes |
| 17 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2183 | 0.115 | 0.6821 | Yes |
| 18 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 2203 | 0.114 | 0.6967 | Yes |
| 19 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.6579 | No |
| 20 | COASY | COASY Entrez, Source | Coenzyme A synthase | 3537 | 0.070 | 0.6188 | No |
| 21 | GLRX2 | GLRX2 Entrez, Source | glutaredoxin 2 | 3592 | 0.069 | 0.6244 | No |
| 22 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.6102 | No |
| 23 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.5833 | No |
| 24 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 4973 | 0.038 | 0.5422 | No |
| 25 | TSPO | TSPO Entrez, Source | translocator protein (18kDa) | 5475 | 0.028 | 0.5085 | No |
| 26 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 6525 | 0.007 | 0.4307 | No |
| 27 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7856 | -0.020 | 0.3336 | No |
| 28 | CPOX | CPOX Entrez, Source | coproporphyrinogen oxidase | 8432 | -0.033 | 0.2950 | No |
| 29 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 8939 | -0.045 | 0.2632 | No |
| 30 | PPT1 | PPT1 Entrez, Source | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 9878 | -0.071 | 0.2026 | No |
| 31 | GPX1 | GPX1 Entrez, Source | glutathione peroxidase 1 | 11620 | -0.145 | 0.0921 | No |
| 32 | COX15 | COX15 Entrez, Source | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 12707 | -0.266 | 0.0477 | No |