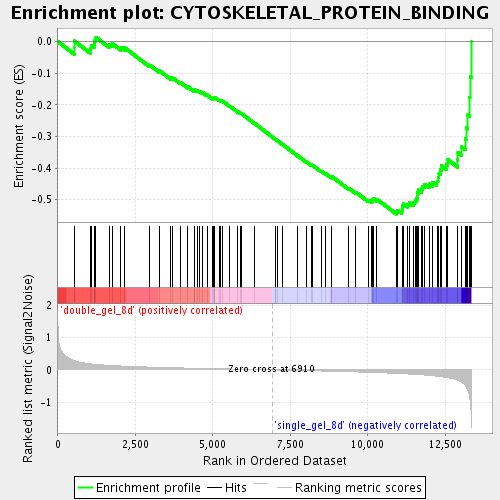
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | CYTOSKELETAL_PROTEIN_BINDING |
| Enrichment Score (ES) | -0.54565877 |
| Normalized Enrichment Score (NES) | -1.932388 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.017669238 |
| FWER p-Value | 0.322 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ANK1 | ANK1 Entrez, Source | ankyrin 1, erythrocytic | 525 | 0.292 | -0.0186 | No |
| 2 | NCALD | NCALD Entrez, Source | neurocalcin delta | 527 | 0.292 | 0.0023 | No |
| 3 | TNNT1 | TNNT1 Entrez, Source | troponin T type 1 (skeletal, slow) | 1040 | 0.188 | -0.0228 | No |
| 4 | GC | GC Entrez, Source | group-specific component (vitamin D binding protein) | 1067 | 0.186 | -0.0114 | No |
| 5 | SYNE1 | SYNE1 Entrez, Source | spectrin repeat containing, nuclear envelope 1 | 1170 | 0.174 | -0.0066 | No |
| 6 | ABRA | ABRA Entrez, Source | actin-binding Rho activating protein | 1190 | 0.172 | 0.0043 | No |
| 7 | UXT | UXT Entrez, Source | ubiquitously-expressed transcript | 1225 | 0.169 | 0.0139 | No |
| 8 | SPTBN4 | SPTBN4 Entrez, Source | spectrin, beta, non-erythrocytic 4 | 1651 | 0.140 | -0.0081 | No |
| 9 | TNNC1 | TNNC1 Entrez, Source | troponin C type 1 (slow) | 1752 | 0.134 | -0.0061 | No |
| 10 | LRPPRC | LRPPRC Entrez, Source | leucine-rich PPR-motif containing | 2030 | 0.121 | -0.0183 | No |
| 11 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 2146 | 0.116 | -0.0186 | No |
| 12 | HPCA | HPCA Entrez, Source | hippocalcin | 2962 | 0.086 | -0.0739 | No |
| 13 | NRCAM | NRCAM Entrez, Source | neuronal cell adhesion molecule | 3263 | 0.077 | -0.0910 | No |
| 14 | MAPK8IP2 | MAPK8IP2 Entrez, Source | mitogen-activated protein kinase 8 interacting protein 2 | 3638 | 0.068 | -0.1143 | No |
| 15 | DYNC1I1 | DYNC1I1 Entrez, Source | dynein, cytoplasmic 1, intermediate chain 1 | 3708 | 0.066 | -0.1148 | No |
| 16 | SCIN | SCIN Entrez, Source | scinderin | 3942 | 0.061 | -0.1279 | No |
| 17 | TNNI3 | TNNI3 Entrez, Source | troponin I type 3 (cardiac) | 4196 | 0.055 | -0.1431 | No |
| 18 | TTN | TTN Entrez, Source | titin | 4401 | 0.050 | -0.1548 | No |
| 19 | ARPC1A | ARPC1A Entrez, Source | actin related protein 2/3 complex, subunit 1A, 41kDa | 4419 | 0.050 | -0.1525 | No |
| 20 | HOOK3 | HOOK3 Entrez, Source | hook homolog 3 (Drosophila) | 4489 | 0.049 | -0.1542 | No |
| 21 | TNNT2 | TNNT2 Entrez, Source | troponin T type 2 (cardiac) | 4572 | 0.047 | -0.1570 | No |
| 22 | RPH3AL | RPH3AL Entrez, Source | rabphilin 3A-like (without C2 domains) | 4660 | 0.045 | -0.1604 | No |
| 23 | MAPT | MAPT Entrez, Source | microtubule-associated protein tau | 4812 | 0.042 | -0.1688 | No |
| 24 | DCX | DCX Entrez, Source | doublecortex; lissencephaly, X-linked (doublecortin) | 5004 | 0.038 | -0.1804 | No |
| 25 | RHCG | RHCG Entrez, Source | Rh family, C glycoprotein | 5006 | 0.038 | -0.1778 | No |
| 26 | CENPJ | CENPJ Entrez, Source | centromere protein J | 5047 | 0.037 | -0.1781 | No |
| 27 | APOE | APOE Entrez, Source | apolipoprotein E | 5056 | 0.037 | -0.1761 | No |
| 28 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 5204 | 0.034 | -0.1847 | No |
| 29 | ADD2 | ADD2 Entrez, Source | adducin 2 (beta) | 5257 | 0.033 | -0.1863 | No |
| 30 | GABARAPL2 | GABARAPL2 Entrez, Source | GABA(A) receptor-associated protein-like 2 | 5307 | 0.032 | -0.1877 | No |
| 31 | VAPB | VAPB Entrez, Source | VAMP (vesicle-associated membrane protein)-associated protein B and C | 5545 | 0.027 | -0.2037 | No |
| 32 | CCR5 | CCR5 Entrez, Source | chemokine (C-C motif) receptor 5 | 5806 | 0.022 | -0.2217 | No |
| 33 | RHAG | RHAG Entrez, Source | Rh-associated glycoprotein | 5878 | 0.021 | -0.2256 | No |
| 34 | MAPK8IP3 | MAPK8IP3 Entrez, Source | mitogen-activated protein kinase 8 interacting protein 3 | 5918 | 0.020 | -0.2271 | No |
| 35 | TRIM63 | TRIM63 Entrez, Source | tripartite motif-containing 63 | 6351 | 0.011 | -0.2589 | No |
| 36 | MYBPC1 | MYBPC1 Entrez, Source | myosin binding protein C, slow type | 7017 | -0.002 | -0.3089 | No |
| 37 | MEFV | MEFV Entrez, Source | Mediterranean fever | 7026 | -0.003 | -0.3093 | No |
| 38 | GABARAP | GABARAP Entrez, Source | GABA(A) receptor-associated protein | 7091 | -0.004 | -0.3139 | No |
| 39 | WASF2 | WASF2 Entrez, Source | WAS protein family, member 2 | 7261 | -0.008 | -0.3261 | No |
| 40 | KATNA1 | KATNA1 Entrez, Source | katanin p60 (ATPase-containing) subunit A 1 | 7746 | -0.018 | -0.3613 | No |
| 41 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 8008 | -0.024 | -0.3793 | No |
| 42 | KIF1B | KIF1B Entrez, Source | kinesin family member 1B | 8176 | -0.027 | -0.3900 | No |
| 43 | SYNPO | SYNPO Entrez, Source | synaptopodin | 8210 | -0.028 | -0.3905 | No |
| 44 | ESPN | ESPN Entrez, Source | espin | 8501 | -0.034 | -0.4099 | No |
| 45 | SORBS3 | SORBS3 Entrez, Source | sorbin and SH3 domain containing 3 | 8634 | -0.037 | -0.4171 | No |
| 46 | TMOD2 | TMOD2 Entrez, Source | tropomodulin 2 (neuronal) | 8827 | -0.042 | -0.4286 | No |
| 47 | RAE1 | RAE1 Entrez, Source | RAE1 RNA export 1 homolog (S. pombe) | 8844 | -0.042 | -0.4268 | No |
| 48 | MARCKS | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 9387 | -0.057 | -0.4636 | No |
| 49 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 9617 | -0.063 | -0.4764 | No |
| 50 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 10026 | -0.075 | -0.5017 | No |
| 51 | DBNL | DBNL Entrez, Source | drebrin-like | 10114 | -0.079 | -0.5026 | No |
| 52 | MYO9B | MYO9B Entrez, Source | myosin IXB | 10160 | -0.080 | -0.5003 | No |
| 53 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 10171 | -0.080 | -0.4952 | No |
| 54 | APC | APC Entrez, Source | adenomatosis polyposis coli | 10296 | -0.085 | -0.4985 | No |
| 55 | CAPZB | CAPZB Entrez, Source | capping protein (actin filament) muscle Z-line, beta | 10922 | -0.108 | -0.5379 | Yes |
| 56 | TMOD3 | TMOD3 Entrez, Source | tropomodulin 3 (ubiquitous) | 10964 | -0.110 | -0.5331 | Yes |
| 57 | ARHGEF2 | ARHGEF2 Entrez, Source | rho/rac guanine nucleotide exchange factor (GEF) 2 | 11105 | -0.117 | -0.5352 | Yes |
| 58 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 11117 | -0.118 | -0.5276 | Yes |
| 59 | TLN1 | TLN1 Entrez, Source | talin 1 | 11127 | -0.118 | -0.5198 | Yes |
| 60 | LSP1 | LSP1 Entrez, Source | lymphocyte-specific protein 1 | 11156 | -0.119 | -0.5133 | Yes |
| 61 | POLB | POLB Entrez, Source | polymerase (DNA directed), beta | 11294 | -0.126 | -0.5145 | Yes |
| 62 | PACSIN3 | PACSIN3 Entrez, Source | protein kinase C and casein kinase substrate in neurons 3 | 11342 | -0.128 | -0.5089 | Yes |
| 63 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 11484 | -0.136 | -0.5097 | Yes |
| 64 | SMTN | SMTN Entrez, Source | smoothelin | 11525 | -0.139 | -0.5027 | Yes |
| 65 | BAIAP2 | BAIAP2 Entrez, Source | BAI1-associated protein 2 | 11565 | -0.141 | -0.4955 | Yes |
| 66 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 11603 | -0.144 | -0.4880 | Yes |
| 67 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 11608 | -0.144 | -0.4779 | Yes |
| 68 | CALD1 | CALD1 Entrez, Source | caldesmon 1 | 11630 | -0.145 | -0.4690 | Yes |
| 69 | S100B | S100B Entrez, Source | S100 calcium binding protein B | 11736 | -0.151 | -0.4661 | Yes |
| 70 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 11762 | -0.153 | -0.4569 | Yes |
| 71 | PXN | PXN Entrez, Source | paxillin | 11831 | -0.159 | -0.4506 | Yes |
| 72 | CDK5RAP2 | CDK5RAP2 Entrez, Source | CDK5 regulatory subunit associated protein 2 | 11990 | -0.172 | -0.4502 | Yes |
| 73 | ABI1 | ABI1 Entrez, Source | abl-interactor 1 | 12095 | -0.181 | -0.4450 | Yes |
| 74 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 12234 | -0.197 | -0.4412 | Yes |
| 75 | ACTN4 | ACTN4 Entrez, Source | actinin, alpha 4 | 12269 | -0.201 | -0.4293 | Yes |
| 76 | SORBS2 | SORBS2 Entrez, Source | sorbin and SH3 domain containing 2 | 12297 | -0.204 | -0.4167 | Yes |
| 77 | JUP | JUP Entrez, Source | junction plakoglobin | 12351 | -0.210 | -0.4056 | Yes |
| 78 | PRNP | PRNP Entrez, Source | prion protein (p27-30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 12376 | -0.212 | -0.3921 | Yes |
| 79 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 12533 | -0.233 | -0.3871 | Yes |
| 80 | MYH9 | MYH9 Entrez, Source | myosin, heavy chain 9, non-muscle | 12585 | -0.241 | -0.3736 | Yes |
| 81 | ADD1 | ADD1 Entrez, Source | adducin 1 (alpha) | 12900 | -0.322 | -0.3741 | Yes |
| 82 | CLASP1 | CLASP1 Entrez, Source | cytoplasmic linker associated protein 1 | 12909 | -0.326 | -0.3513 | Yes |
| 83 | NEXN | NEXN Entrez, Source | nexilin (F actin binding protein) | 13033 | -0.382 | -0.3331 | Yes |
| 84 | FXYD5 | FXYD5 Entrez, Source | FXYD domain containing ion transport regulator 5 | 13147 | -0.472 | -0.3076 | Yes |
| 85 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 13173 | -0.513 | -0.2726 | Yes |
| 86 | ACTN1 | ACTN1 Entrez, Source | actinin, alpha 1 | 13226 | -0.619 | -0.2320 | Yes |
| 87 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13288 | -0.823 | -0.1774 | Yes |
| 88 | ABI2 | ABI2 Entrez, Source | abl interactor 2 | 13301 | -0.939 | -0.1107 | Yes |
| 89 | ACTA1 | ACTA1 Entrez, Source | actin, alpha 1, skeletal muscle | 13340 | -1.581 | 0.0002 | Yes |