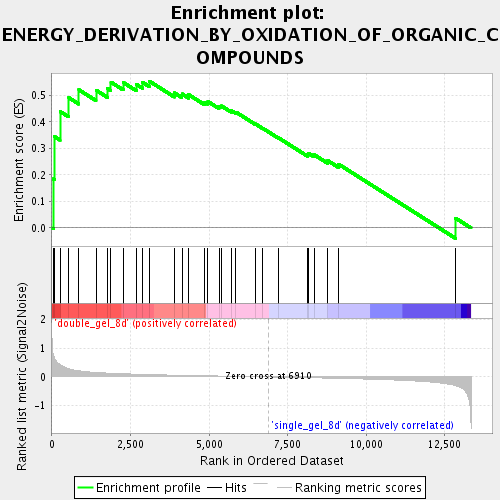
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | ENERGY_DERIVATION_BY_OXIDATION_OF_ORGANIC_COMPOUNDS |
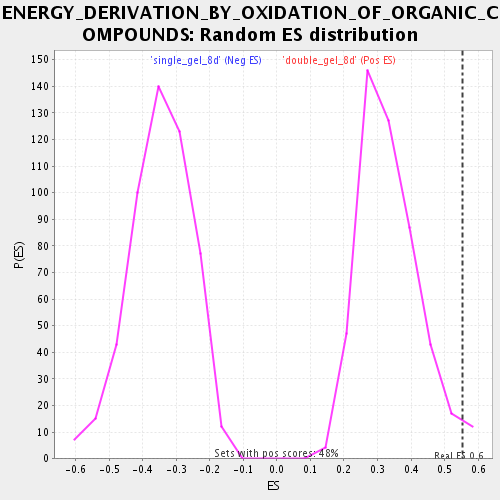
| Enrichment Score (ES) | 0.5533666 |
| Normalized Enrichment Score (NES) | 1.6440405 |
| Nominal p-value | 0.022774328 |
| FDR q-value | 0.051555928 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GYS2 | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 52 | 0.748 | 0.1850 | Yes |
| 2 | LEPR | LEPR Entrez, Source | leptin receptor | 85 | 0.639 | 0.3440 | Yes |
| 3 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.4390 | Yes |
| 4 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 537 | 0.288 | 0.4909 | Yes |
| 5 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.5205 | Yes |
| 6 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.5192 | Yes |
| 7 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1770 | 0.133 | 0.5253 | Yes |
| 8 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 1880 | 0.127 | 0.5493 | Yes |
| 9 | BCS1L | BCS1L Entrez, Source | BCS1-like (yeast) | 2272 | 0.111 | 0.5480 | Yes |
| 10 | EPM2A | EPM2A Entrez, Source | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 2687 | 0.096 | 0.5411 | Yes |
| 11 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.5486 | Yes |
| 12 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 3098 | 0.082 | 0.5534 | Yes |
| 13 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.5090 | No |
| 14 | UQCRC2 | UQCRC2 Entrez, Source | ubiquinol-cytochrome c reductase core protein II | 4141 | 0.056 | 0.5051 | No |
| 15 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.5022 | No |
| 16 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 4861 | 0.041 | 0.4744 | No |
| 17 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 4966 | 0.039 | 0.4763 | No |
| 18 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 5320 | 0.032 | 0.4578 | No |
| 19 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5395 | 0.030 | 0.4599 | No |
| 20 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 5716 | 0.023 | 0.4417 | No |
| 21 | ADRB3 | ADRB3 Entrez, Source | adrenergic, beta-3-, receptor | 5857 | 0.021 | 0.4365 | No |
| 22 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 6477 | 0.008 | 0.3921 | No |
| 23 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 6698 | 0.004 | 0.3766 | No |
| 24 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 7209 | -0.006 | 0.3399 | No |
| 25 | LEP | LEP Entrez, Source | leptin (obesity homolog, mouse) | 8144 | -0.026 | 0.2764 | No |
| 26 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 8170 | -0.027 | 0.2813 | No |
| 27 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 8354 | -0.031 | 0.2755 | No |
| 28 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 8782 | -0.040 | 0.2536 | No |
| 29 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 9121 | -0.049 | 0.2407 | No |
| 30 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 12849 | -0.302 | 0.0370 | No |