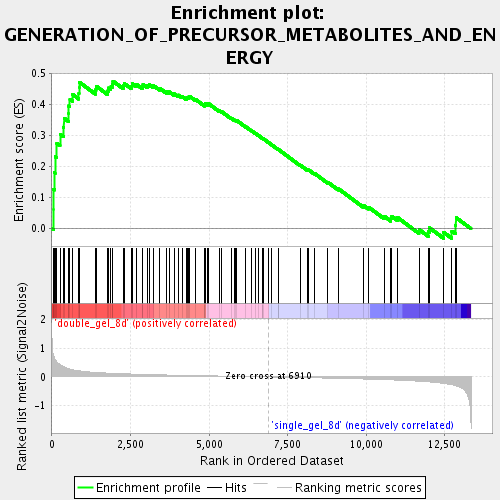
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | GENERATION_OF_PRECURSOR_METABOLITES_AND_ENERGY |
| Enrichment Score (ES) | 0.47539312 |
| Normalized Enrichment Score (NES) | 1.7471393 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.021617595 |
| FWER p-Value | 0.981 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 50 | 0.751 | 0.0618 | Yes |
| 2 | GYS2 | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 52 | 0.748 | 0.1270 | Yes |
| 3 | LEPR | LEPR Entrez, Source | leptin receptor | 85 | 0.639 | 0.1804 | Yes |
| 4 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 104 | 0.612 | 0.2325 | Yes |
| 5 | APOM | APOM Entrez, Source | apolipoprotein M | 156 | 0.527 | 0.2746 | Yes |
| 6 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.3043 | Yes |
| 7 | GBE1 | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 368 | 0.355 | 0.3271 | Yes |
| 8 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 384 | 0.346 | 0.3563 | Yes |
| 9 | GCGR | GCGR Entrez, Source | glucagon receptor | 531 | 0.290 | 0.3705 | Yes |
| 10 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 537 | 0.288 | 0.3953 | Yes |
| 11 | AQP7 | AQP7 Entrez, Source | aquaporin 7 | 573 | 0.276 | 0.4168 | Yes |
| 12 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 656 | 0.251 | 0.4325 | Yes |
| 13 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.4359 | Yes |
| 14 | PDIA5 | PDIA5 Entrez, Source | protein disulfide isomerase family A, member 5 | 864 | 0.212 | 0.4541 | Yes |
| 15 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 870 | 0.211 | 0.4721 | Yes |
| 16 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 1386 | 0.158 | 0.4471 | Yes |
| 17 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.4595 | Yes |
| 18 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1770 | 0.133 | 0.4436 | Yes |
| 19 | AVP | AVP Entrez, Source | arginine vasopressin (neurophysin II, antidiuretic hormone, diabetes insipidus, neurohypophyseal) | 1794 | 0.131 | 0.4533 | Yes |
| 20 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 1880 | 0.127 | 0.4580 | Yes |
| 21 | COX17 | COX17 Entrez, Source | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) | 1921 | 0.125 | 0.4660 | Yes |
| 22 | DHRS4 | DHRS4 Entrez, Source | dehydrogenase/reductase (SDR family) member 4 | 1941 | 0.124 | 0.4754 | Yes |
| 23 | BCS1L | BCS1L Entrez, Source | BCS1-like (yeast) | 2272 | 0.111 | 0.4602 | No |
| 24 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 2294 | 0.111 | 0.4683 | No |
| 25 | PPARG | PPARG Entrez, Source | peroxisome proliferative activated receptor, gamma | 2538 | 0.101 | 0.4588 | No |
| 26 | SPR | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 2551 | 0.101 | 0.4668 | No |
| 27 | EPM2A | EPM2A Entrez, Source | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 2687 | 0.096 | 0.4650 | No |
| 28 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.4577 | No |
| 29 | INSR | INSR Entrez, Source | insulin receptor | 2897 | 0.088 | 0.4647 | No |
| 30 | PHKG2 | PHKG2 Entrez, Source | phosphorylase kinase, gamma 2 (testis) | 3036 | 0.084 | 0.4616 | No |
| 31 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 3098 | 0.082 | 0.4641 | No |
| 32 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 3231 | 0.078 | 0.4609 | No |
| 33 | CYB5R2 | CYB5R2 Entrez, Source | cytochrome b5 reductase 2 | 3428 | 0.073 | 0.4525 | No |
| 34 | MTRR | MTRR Entrez, Source | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 3648 | 0.068 | 0.4419 | No |
| 35 | QPRT | QPRT Entrez, Source | quinolinate phosphoribosyltransferase (nicotinate-nucleotide pyrophosphorylase (carboxylating)) | 3739 | 0.065 | 0.4409 | No |
| 36 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.4344 | No |
| 37 | PHKB | PHKB Entrez, Source | phosphorylase kinase, beta | 4016 | 0.059 | 0.4307 | No |
| 38 | UQCRC2 | UQCRC2 Entrez, Source | ubiquinol-cytochrome c reductase core protein II | 4141 | 0.056 | 0.4263 | No |
| 39 | POMC | POMC Entrez, Source | proopiomelanocortin (adrenocorticotropin/ beta-lipotropin/ alpha-melanocyte stimulating hormone/ beta-melanocyte stimulating hormone/ beta-endorphin) | 4293 | 0.053 | 0.4195 | No |
| 40 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 4305 | 0.053 | 0.4233 | No |
| 41 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.4242 | No |
| 42 | XYLB | XYLB Entrez, Source | xylulokinase homolog (H. influenzae) | 4379 | 0.051 | 0.4268 | No |
| 43 | GIPR | GIPR Entrez, Source | gastric inhibitory polypeptide receptor | 4561 | 0.047 | 0.4172 | No |
| 44 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 4861 | 0.041 | 0.3982 | No |
| 45 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 4870 | 0.040 | 0.4012 | No |
| 46 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 4884 | 0.040 | 0.4037 | No |
| 47 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 4966 | 0.039 | 0.4009 | No |
| 48 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 4973 | 0.038 | 0.4038 | No |
| 49 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 5320 | 0.032 | 0.3805 | No |
| 50 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5395 | 0.030 | 0.3776 | No |
| 51 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 5716 | 0.023 | 0.3555 | No |
| 52 | ISL1 | ISL1 Entrez, Source | ISL1 transcription factor, LIM/homeodomain, (islet-1) | 5811 | 0.022 | 0.3503 | No |
| 53 | ADRB3 | ADRB3 Entrez, Source | adrenergic, beta-3-, receptor | 5857 | 0.021 | 0.3487 | No |
| 54 | WFS1 | WFS1 Entrez, Source | Wolfram syndrome 1 (wolframin) | 5888 | 0.020 | 0.3483 | No |
| 55 | SLC25A27 | SLC25A27 Entrez, Source | solute carrier family 25, member 27 | 6177 | 0.015 | 0.3278 | No |
| 56 | DUOX2 | DUOX2 Entrez, Source | dual oxidase 2 | 6346 | 0.011 | 0.3161 | No |
| 57 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 6477 | 0.008 | 0.3070 | No |
| 58 | ADIPOQ | ADIPOQ Entrez, Source | adiponectin, C1Q and collagen domain containing | 6570 | 0.007 | 0.3006 | No |
| 59 | NOX4 | NOX4 Entrez, Source | NADPH oxidase 4 | 6694 | 0.004 | 0.2917 | No |
| 60 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 6698 | 0.004 | 0.2919 | No |
| 61 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 6721 | 0.004 | 0.2905 | No |
| 62 | RETSAT | RETSAT Entrez, Source | retinol saturase (all-trans-retinol 13,14-reductase) | 6880 | 0.001 | 0.2787 | No |
| 63 | MCHR1 | MCHR1 Entrez, Source | melanin-concentrating hormone receptor 1 | 7002 | -0.002 | 0.2697 | No |
| 64 | TXNL1 | TXNL1 Entrez, Source | thioredoxin-like 1 | 7208 | -0.006 | 0.2548 | No |
| 65 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 7209 | -0.006 | 0.2554 | No |
| 66 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 7908 | -0.021 | 0.2046 | No |
| 67 | LEP | LEP Entrez, Source | leptin (obesity homolog, mouse) | 8144 | -0.026 | 0.1892 | No |
| 68 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 8170 | -0.027 | 0.1897 | No |
| 69 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 8354 | -0.031 | 0.1786 | No |
| 70 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 8782 | -0.040 | 0.1499 | No |
| 71 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 9121 | -0.049 | 0.1287 | No |
| 72 | ENPP1 | ENPP1 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 9921 | -0.072 | 0.0748 | No |
| 73 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 10075 | -0.077 | 0.0700 | No |
| 74 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 10578 | -0.094 | 0.0403 | No |
| 75 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 10791 | -0.102 | 0.0333 | No |
| 76 | ATPIF1 | ATPIF1 Entrez, Source | ATPase inhibitory factor 1 | 10815 | -0.103 | 0.0406 | No |
| 77 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 11000 | -0.112 | 0.0364 | No |
| 78 | PHKA1 | PHKA1 Entrez, Source | phosphorylase kinase, alpha 1 (muscle) | 11687 | -0.149 | -0.0023 | No |
| 79 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 11983 | -0.171 | -0.0096 | No |
| 80 | AVPR1A | AVPR1A Entrez, Source | arginine vasopressin receptor 1A | 12016 | -0.174 | 0.0032 | No |
| 81 | CROT | CROT Entrez, Source | carnitine O-octanoyltransferase | 12469 | -0.223 | -0.0114 | No |
| 82 | PPP1R2 | PPP1R2 Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 2 | 12730 | -0.272 | -0.0073 | No |
| 83 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 12849 | -0.302 | 0.0102 | No |
| 84 | LOXL1 | LOXL1 Entrez, Source | lysyl oxidase-like 1 | 12864 | -0.308 | 0.0361 | No |