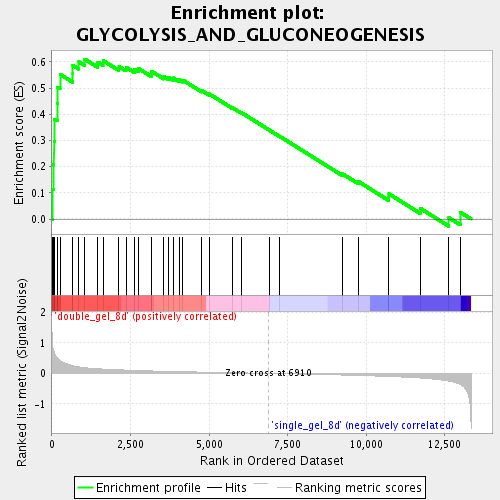
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | 0.61041373 |
| Normalized Enrichment Score (NES) | 1.8897561 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0060452954 |
| FWER p-Value | 0.503 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 21 | 0.893 | 0.1144 | Yes |
| 2 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 56 | 0.731 | 0.2068 | Yes |
| 3 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 66 | 0.702 | 0.2974 | Yes |
| 4 | PC | PC Entrez, Source | pyruvate carboxylase | 79 | 0.652 | 0.3813 | Yes |
| 5 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 167 | 0.505 | 0.4405 | Yes |
| 6 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 184 | 0.487 | 0.5026 | Yes |
| 7 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.5526 | Yes |
| 8 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 663 | 0.250 | 0.5547 | Yes |
| 9 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 666 | 0.249 | 0.5869 | Yes |
| 10 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.6002 | Yes |
| 11 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 1047 | 0.187 | 0.6104 | Yes |
| 12 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 1462 | 0.152 | 0.5991 | No |
| 13 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 1628 | 0.141 | 0.6050 | No |
| 14 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 2134 | 0.117 | 0.5822 | No |
| 15 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2361 | 0.108 | 0.5793 | No |
| 16 | ENO2 | ENO2 Entrez, Source | enolase 2 (gamma, neuronal) | 2627 | 0.098 | 0.5722 | No |
| 17 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 2767 | 0.093 | 0.5738 | No |
| 18 | TPI1 | TPI1 Entrez, Source | triosephosphate isomerase 1 | 3163 | 0.080 | 0.5545 | No |
| 19 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 3168 | 0.079 | 0.5645 | No |
| 20 | PGK1 | PGK1 Entrez, Source | phosphoglycerate kinase 1 | 3566 | 0.069 | 0.5437 | No |
| 21 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 3707 | 0.066 | 0.5418 | No |
| 22 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 3857 | 0.063 | 0.5387 | No |
| 23 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 4054 | 0.058 | 0.5316 | No |
| 24 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 4168 | 0.056 | 0.5303 | No |
| 25 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 4772 | 0.042 | 0.4905 | No |
| 26 | PGAM2 | PGAM2 Entrez, Source | phosphoglycerate mutase 2 (muscle) | 5008 | 0.038 | 0.4778 | No |
| 27 | GAPDH | GAPDH Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase | 5744 | 0.023 | 0.4255 | No |
| 28 | ENO3 | ENO3 Entrez, Source | enolase 3 (beta, muscle) | 6027 | 0.018 | 0.4066 | No |
| 29 | HK2 | HK2 Entrez, Source | hexokinase 2 | 6922 | -0.000 | 0.3395 | No |
| 30 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 7234 | -0.007 | 0.3170 | No |
| 31 | PGAM1 | PGAM1 Entrez, Source | phosphoglycerate mutase 1 (brain) | 9249 | -0.053 | 0.1725 | No |
| 32 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9745 | -0.066 | 0.1440 | No |
| 33 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 10723 | -0.099 | 0.0834 | No |
| 34 | LDHAL6B | LDHAL6B Entrez, Source | lactate dehydrogenase A-like 6B | 10728 | -0.099 | 0.0961 | No |
| 35 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 11723 | -0.150 | 0.0409 | No |
| 36 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12636 | -0.251 | 0.0050 | No |
| 37 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 13007 | -0.369 | 0.0252 | No |