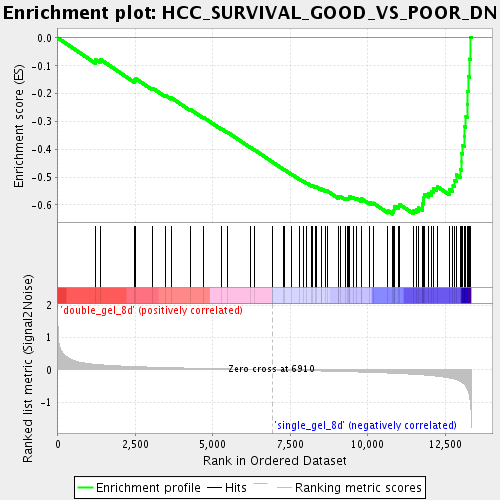
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | HCC_SURVIVAL_GOOD_VS_POOR_DN |
| Enrichment Score (ES) | -0.63286614 |
| Normalized Enrichment Score (NES) | -2.1760921 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0010482055 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 1205 | 0.170 | -0.0773 | No |
| 2 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 1363 | 0.160 | -0.0764 | No |
| 3 | LYAR | LYAR Entrez, Source | - | 2462 | 0.104 | -0.1508 | No |
| 4 | RPL36A | RPL36A Entrez, Source | ribosomal protein L36a | 2516 | 0.102 | -0.1467 | No |
| 5 | PELI1 | PELI1 Entrez, Source | pellino homolog 1 (Drosophila) | 3050 | 0.083 | -0.1802 | No |
| 6 | RWDD1 | RWDD1 Entrez, Source | RWD domain containing 1 | 3474 | 0.072 | -0.2064 | No |
| 7 | IER3 | IER3 Entrez, Source | immediate early response 3 | 3649 | 0.068 | -0.2141 | No |
| 8 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 4283 | 0.053 | -0.2576 | No |
| 9 | SLC34A1 | SLC34A1 Entrez, Source | solute carrier family 34 (sodium phosphate), member 1 | 4690 | 0.044 | -0.2846 | No |
| 10 | CD164 | CD164 Entrez, Source | CD164 molecule, sialomucin | 5266 | 0.032 | -0.3254 | No |
| 11 | RALA | RALA Entrez, Source | v-ral simian leukemia viral oncogene homolog A (ras related) | 5473 | 0.028 | -0.3387 | No |
| 12 | MYH2 | MYH2 Entrez, Source | myosin, heavy chain 2, skeletal muscle, adult | 6204 | 0.014 | -0.3926 | No |
| 13 | NSF | NSF Entrez, Source | N-ethylmaleimide-sensitive factor | 6347 | 0.011 | -0.4024 | No |
| 14 | NAP1L1 | NAP1L1 Entrez, Source | nucleosome assembly protein 1-like 1 | 6915 | -0.000 | -0.4452 | No |
| 15 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 6925 | -0.000 | -0.4458 | No |
| 16 | RPL35 | RPL35 Entrez, Source | ribosomal protein L35 | 7279 | -0.008 | -0.4718 | No |
| 17 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 7299 | -0.008 | -0.4726 | No |
| 18 | RPS18 | RPS18 Entrez, Source | ribosomal protein S18 | 7553 | -0.014 | -0.4906 | No |
| 19 | RPS6 | RPS6 Entrez, Source | ribosomal protein S6 | 7800 | -0.019 | -0.5076 | No |
| 20 | TES | TES Entrez, Source | testis derived transcript (3 LIM domains) | 7914 | -0.021 | -0.5144 | No |
| 21 | OPLAH | OPLAH Entrez, Source | 5-oxoprolinase (ATP-hydrolysing) | 8035 | -0.024 | -0.5215 | No |
| 22 | RPL31 | RPL31 Entrez, Source | ribosomal protein L31 | 8192 | -0.027 | -0.5311 | No |
| 23 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 8204 | -0.028 | -0.5297 | No |
| 24 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 8325 | -0.031 | -0.5363 | No |
| 25 | RPL9 | RPL9 Entrez, Source | ribosomal protein L9 | 8332 | -0.031 | -0.5343 | No |
| 26 | ROD1 | ROD1 Entrez, Source | ROD1 regulator of differentiation 1 (S. pombe) | 8498 | -0.034 | -0.5440 | No |
| 27 | RPS3A | RPS3A Entrez, Source | ribosomal protein S3A | 8514 | -0.035 | -0.5424 | No |
| 28 | RPL17 | RPL17 Entrez, Source | ribosomal protein L17 | 8625 | -0.037 | -0.5477 | No |
| 29 | RPS9 | RPS9 Entrez, Source | ribosomal protein S9 | 8689 | -0.039 | -0.5494 | No |
| 30 | HNRPC | HNRPC Entrez, Source | heterogeneous nuclear ribonucleoprotein C (C1/C2) | 9050 | -0.047 | -0.5727 | No |
| 31 | RPL27 | RPL27 Entrez, Source | ribosomal protein L27 | 9053 | -0.047 | -0.5691 | No |
| 32 | CCT6A | CCT6A Entrez, Source | chaperonin containing TCP1, subunit 6A (zeta 1) | 9113 | -0.049 | -0.5696 | No |
| 33 | USP1 | USP1 Entrez, Source | ubiquitin specific peptidase 1 | 9270 | -0.054 | -0.5771 | No |
| 34 | DUSP18 | DUSP18 Entrez, Source | dual specificity phosphatase 18 | 9332 | -0.055 | -0.5773 | No |
| 35 | MARCKS | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 9387 | -0.057 | -0.5769 | No |
| 36 | RPS3 | RPS3 Entrez, Source | ribosomal protein S3 | 9405 | -0.057 | -0.5736 | No |
| 37 | RBX1 | RBX1 Entrez, Source | ring-box 1 | 9418 | -0.058 | -0.5699 | No |
| 38 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 9534 | -0.061 | -0.5737 | No |
| 39 | RPS5 | RPS5 Entrez, Source | ribosomal protein S5 | 9640 | -0.064 | -0.5765 | No |
| 40 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 9794 | -0.068 | -0.5827 | No |
| 41 | HMGN1 | HMGN1 Entrez, Source | high-mobility group nucleosome binding domain 1 | 9807 | -0.069 | -0.5781 | No |
| 42 | LXN | LXN Entrez, Source | latexin | 10057 | -0.076 | -0.5908 | No |
| 43 | FBL | FBL Entrez, Source | fibrillarin | 10172 | -0.080 | -0.5930 | No |
| 44 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 10651 | -0.097 | -0.6213 | No |
| 45 | TMSB4X | TMSB4X Entrez, Source | thymosin, beta 4, X-linked | 10806 | -0.103 | -0.6247 | Yes |
| 46 | CCT5 | CCT5 Entrez, Source | chaperonin containing TCP1, subunit 5 (epsilon) | 10843 | -0.105 | -0.6190 | Yes |
| 47 | NOL5A | NOL5A Entrez, Source | nucleolar protein 5A (56kDa with KKE/D repeat) | 10849 | -0.105 | -0.6110 | Yes |
| 48 | BZW2 | BZW2 Entrez, Source | basic leucine zipper and W2 domains 2 | 10870 | -0.106 | -0.6041 | Yes |
| 49 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 10991 | -0.111 | -0.6043 | Yes |
| 50 | ANXA4 | ANXA4 Entrez, Source | annexin A4 | 11026 | -0.113 | -0.5979 | Yes |
| 51 | ITPR3 | ITPR3 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 3 | 11479 | -0.136 | -0.6211 | Yes |
| 52 | ACTR3 | ACTR3 Entrez, Source | ARP3 actin-related protein 3 homolog (yeast) | 11578 | -0.142 | -0.6171 | Yes |
| 53 | CALM2 | CALM2 Entrez, Source | calmodulin 2 (phosphorylase kinase, delta) | 11648 | -0.147 | -0.6106 | Yes |
| 54 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 11762 | -0.153 | -0.6069 | Yes |
| 55 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 11774 | -0.155 | -0.5954 | Yes |
| 56 | CBX3 | CBX3 Entrez, Source | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 11802 | -0.156 | -0.5849 | Yes |
| 57 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 11811 | -0.158 | -0.5730 | Yes |
| 58 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 11816 | -0.158 | -0.5607 | Yes |
| 59 | DEK | DEK Entrez, Source | DEK oncogene (DNA binding) | 11962 | -0.170 | -0.5581 | Yes |
| 60 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 12056 | -0.178 | -0.5509 | Yes |
| 61 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 12130 | -0.184 | -0.5417 | Yes |
| 62 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 12234 | -0.197 | -0.5337 | Yes |
| 63 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 12630 | -0.250 | -0.5436 | Yes |
| 64 | ANXA3 | ANXA3 Entrez, Source | annexin A3 | 12750 | -0.275 | -0.5306 | Yes |
| 65 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 12794 | -0.284 | -0.5112 | Yes |
| 66 | S100A6 | S100A6 Entrez, Source | S100 calcium binding protein A6 | 12854 | -0.305 | -0.4913 | Yes |
| 67 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 12986 | -0.357 | -0.4727 | Yes |
| 68 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13028 | -0.381 | -0.4454 | Yes |
| 69 | CLIC1 | CLIC1 Entrez, Source | chloride intracellular channel 1 | 13030 | -0.381 | -0.4151 | Yes |
| 70 | HN1 | HN1 Entrez, Source | hematological and neurological expressed 1 | 13064 | -0.406 | -0.3852 | Yes |
| 71 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 13120 | -0.443 | -0.3540 | Yes |
| 72 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 13128 | -0.448 | -0.3188 | Yes |
| 73 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13167 | -0.505 | -0.2814 | Yes |
| 74 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 13212 | -0.580 | -0.2385 | Yes |
| 75 | SERPINH1 | SERPINH1 Entrez, Source | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 13222 | -0.606 | -0.1908 | Yes |
| 76 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13247 | -0.677 | -0.1386 | Yes |
| 77 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13289 | -0.828 | -0.0756 | Yes |
| 78 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 13309 | -0.997 | 0.0025 | Yes |

