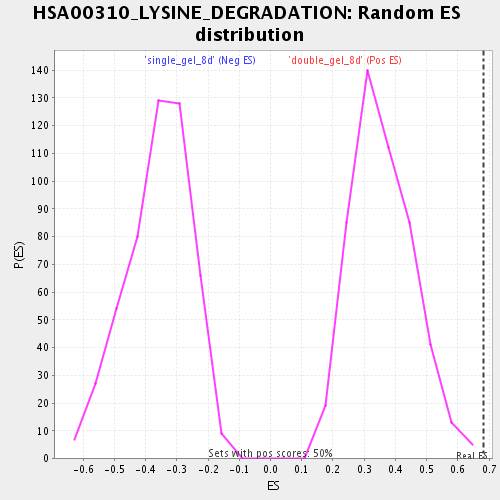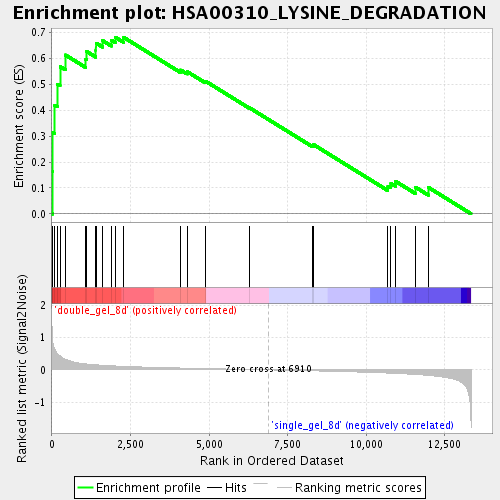
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | HSA00310_LYSINE_DEGRADATION |
| Enrichment Score (ES) | 0.68193 |
| Normalized Enrichment Score (NES) | 1.8961483 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0057881935 |
| FWER p-Value | 0.479 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 16 | 0.933 | 0.1622 | Yes |
| 2 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 24 | 0.864 | 0.3130 | Yes |
| 3 | AADAT | AADAT Entrez, Source | aminoadipate aminotransferase | 88 | 0.633 | 0.4191 | Yes |
| 4 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 172 | 0.499 | 0.5002 | Yes |
| 5 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.5685 | Yes |
| 6 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 430 | 0.329 | 0.6135 | Yes |
| 7 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 1074 | 0.185 | 0.5975 | Yes |
| 8 | SHMT2 | SHMT2 Entrez, Source | serine hydroxymethyltransferase 2 (mitochondrial) | 1105 | 0.181 | 0.6270 | Yes |
| 9 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1399 | 0.157 | 0.6325 | Yes |
| 10 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 1403 | 0.157 | 0.6597 | Yes |
| 11 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 1615 | 0.142 | 0.6687 | Yes |
| 12 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 1892 | 0.127 | 0.6701 | Yes |
| 13 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 2018 | 0.121 | 0.6819 | Yes |
| 14 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 2281 | 0.111 | 0.6817 | No |
| 15 | AKR1B10 | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 4097 | 0.057 | 0.5555 | No |
| 16 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 4305 | 0.053 | 0.5491 | No |
| 17 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 4884 | 0.040 | 0.5128 | No |
| 18 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.4102 | No |
| 19 | TMLHE | TMLHE Entrez, Source | trimethyllysine hydroxylase, epsilon | 8305 | -0.030 | 0.2634 | No |
| 20 | HSD3B7 | HSD3B7 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 8334 | -0.031 | 0.2667 | No |
| 21 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 10696 | -0.098 | 0.1067 | No |
| 22 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 10791 | -0.102 | 0.1175 | No |
| 23 | PLOD3 | PLOD3 Entrez, Source | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 10940 | -0.109 | 0.1255 | No |
| 24 | EHMT2 | EHMT2 Entrez, Source | euchromatic histone-lysine N-methyltransferase 2 | 11568 | -0.141 | 0.1031 | No |
| 25 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 11983 | -0.171 | 0.1020 | No |