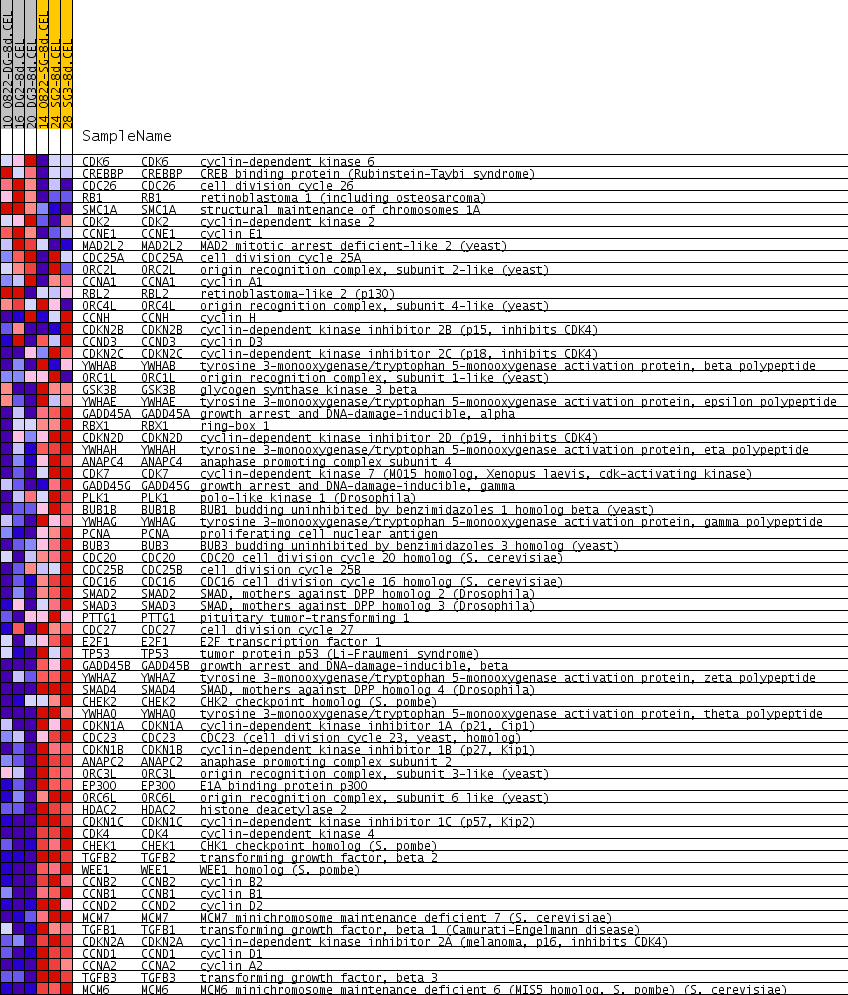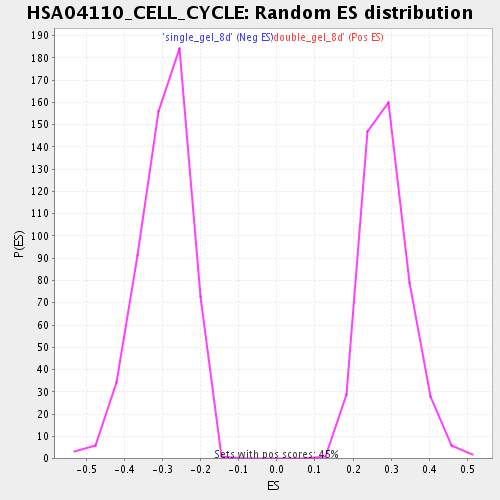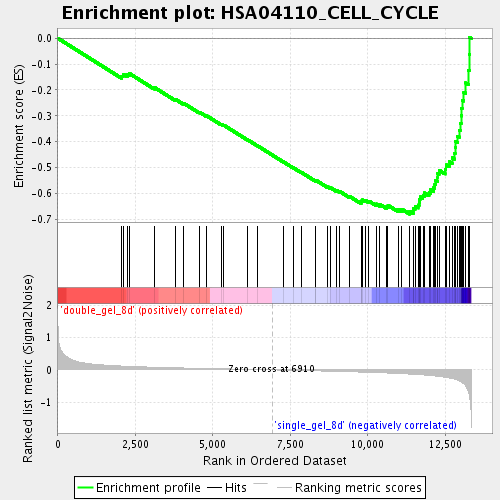
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | HSA04110_CELL_CYCLE |
| Enrichment Score (ES) | -0.68057024 |
| Normalized Enrichment Score (NES) | -2.3058944 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDK6 | CDK6 Entrez, Source | cyclin-dependent kinase 6 | 2067 | 0.119 | -0.1461 | No |
| 2 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 2102 | 0.118 | -0.1391 | No |
| 3 | CDC26 | CDC26 Entrez, Source | cell division cycle 26 | 2240 | 0.113 | -0.1402 | No |
| 4 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 2307 | 0.110 | -0.1363 | No |
| 5 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3109 | 0.081 | -0.1900 | No |
| 6 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 3808 | 0.064 | -0.2374 | No |
| 7 | CCNE1 | CCNE1 Entrez, Source | cyclin E1 | 4056 | 0.058 | -0.2513 | No |
| 8 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 4562 | 0.047 | -0.2855 | No |
| 9 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4801 | 0.042 | -0.3000 | No |
| 10 | ORC2L | ORC2L Entrez, Source | origin recognition complex, subunit 2-like (yeast) | 5273 | 0.032 | -0.3329 | No |
| 11 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 5345 | 0.031 | -0.3357 | No |
| 12 | RBL2 | RBL2 Entrez, Source | retinoblastoma-like 2 (p130) | 6125 | 0.016 | -0.3931 | No |
| 13 | ORC4L | ORC4L Entrez, Source | origin recognition complex, subunit 4-like (yeast) | 6443 | 0.009 | -0.4163 | No |
| 14 | CCNH | CCNH Entrez, Source | cyclin H | 7280 | -0.008 | -0.4786 | No |
| 15 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 7589 | -0.014 | -0.5007 | No |
| 16 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 7859 | -0.020 | -0.5193 | No |
| 17 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 8322 | -0.030 | -0.5516 | No |
| 18 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 8325 | -0.031 | -0.5493 | No |
| 19 | ORC1L | ORC1L Entrez, Source | origin recognition complex, subunit 1-like (yeast) | 8707 | -0.039 | -0.5748 | No |
| 20 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 8782 | -0.040 | -0.5771 | No |
| 21 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8995 | -0.046 | -0.5894 | No |
| 22 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9083 | -0.048 | -0.5920 | No |
| 23 | RBX1 | RBX1 Entrez, Source | ring-box 1 | 9418 | -0.058 | -0.6125 | No |
| 24 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9788 | -0.068 | -0.6348 | No |
| 25 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 9794 | -0.068 | -0.6297 | No |
| 26 | ANAPC4 | ANAPC4 Entrez, Source | anaphase promoting complex subunit 4 | 9814 | -0.069 | -0.6255 | No |
| 27 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9912 | -0.072 | -0.6270 | No |
| 28 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 10034 | -0.076 | -0.6300 | No |
| 29 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 10276 | -0.084 | -0.6413 | No |
| 30 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 10391 | -0.088 | -0.6427 | No |
| 31 | YWHAG | YWHAG Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | 10619 | -0.096 | -0.6521 | No |
| 32 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 10651 | -0.097 | -0.6465 | No |
| 33 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 10991 | -0.111 | -0.6630 | No |
| 34 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11099 | -0.117 | -0.6616 | No |
| 35 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 11352 | -0.129 | -0.6701 | Yes |
| 36 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 11467 | -0.135 | -0.6677 | Yes |
| 37 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 11475 | -0.136 | -0.6572 | Yes |
| 38 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11527 | -0.139 | -0.6498 | Yes |
| 39 | PTTG1 | PTTG1 Entrez, Source | pituitary tumor-transforming 1 | 11624 | -0.145 | -0.6452 | Yes |
| 40 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11658 | -0.148 | -0.6357 | Yes |
| 41 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 11673 | -0.148 | -0.6247 | Yes |
| 42 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11690 | -0.149 | -0.6138 | Yes |
| 43 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 11783 | -0.155 | -0.6081 | Yes |
| 44 | YWHAZ | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 11826 | -0.159 | -0.5984 | Yes |
| 45 | SMAD4 | SMAD4 Entrez, Source | SMAD, mothers against DPP homolog 4 (Drosophila) | 11978 | -0.171 | -0.5959 | Yes |
| 46 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 12031 | -0.176 | -0.5855 | Yes |
| 47 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 12130 | -0.184 | -0.5779 | Yes |
| 48 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 12163 | -0.188 | -0.5651 | Yes |
| 49 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 12182 | -0.190 | -0.5510 | Yes |
| 50 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 12251 | -0.199 | -0.5400 | Yes |
| 51 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 12258 | -0.199 | -0.5243 | Yes |
| 52 | ORC3L | ORC3L Entrez, Source | origin recognition complex, subunit 3-like (yeast) | 12302 | -0.205 | -0.5108 | Yes |
| 53 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 12496 | -0.228 | -0.5068 | Yes |
| 54 | ORC6L | ORC6L Entrez, Source | origin recognition complex, subunit 6 like (yeast) | 12525 | -0.232 | -0.4901 | Yes |
| 55 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 12630 | -0.250 | -0.4776 | Yes |
| 56 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 12725 | -0.270 | -0.4627 | Yes |
| 57 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 12794 | -0.284 | -0.4447 | Yes |
| 58 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12821 | -0.294 | -0.4228 | Yes |
| 59 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12832 | -0.296 | -0.3996 | Yes |
| 60 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 12897 | -0.319 | -0.3784 | Yes |
| 61 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12954 | -0.340 | -0.3550 | Yes |
| 62 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 12986 | -0.357 | -0.3284 | Yes |
| 63 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13016 | -0.374 | -0.3001 | Yes |
| 64 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13028 | -0.381 | -0.2700 | Yes |
| 65 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 13047 | -0.395 | -0.2393 | Yes |
| 66 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 13099 | -0.425 | -0.2086 | Yes |
| 67 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13162 | -0.500 | -0.1727 | Yes |
| 68 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13247 | -0.677 | -0.1240 | Yes |
| 69 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 13277 | -0.783 | -0.0625 | Yes |
| 70 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13289 | -0.828 | 0.0040 | Yes |