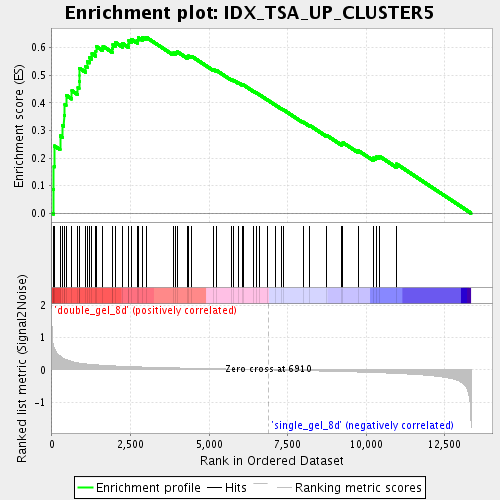
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | IDX_TSA_UP_CLUSTER5 |
| Enrichment Score (ES) | 0.63801265 |
| Normalized Enrichment Score (NES) | 2.1913033 |
| Nominal p-value | 0.0 |
| FDR q-value | 6.8977206E-5 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LPIN1 | LPIN1 Entrez, Source | lipin 1 | 43 | 0.774 | 0.0875 | Yes |
| 2 | PEX11A | PEX11A Entrez, Source | peroxisomal biogenesis factor 11A | 65 | 0.704 | 0.1684 | Yes |
| 3 | PC | PC Entrez, Source | pyruvate carboxylase | 79 | 0.652 | 0.2439 | Yes |
| 4 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.2805 | Yes |
| 5 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 335 | 0.375 | 0.3189 | Yes |
| 6 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 384 | 0.346 | 0.3558 | Yes |
| 7 | TIMM8A | TIMM8A Entrez, Source | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 412 | 0.336 | 0.3931 | Yes |
| 8 | TMEM97 | TMEM97 Entrez, Source | transmembrane protein 97 | 451 | 0.320 | 0.4278 | Yes |
| 9 | C3 | C3 Entrez, Source | complement component 3 | 636 | 0.257 | 0.4440 | Yes |
| 10 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 828 | 0.216 | 0.4550 | Yes |
| 11 | ETFB | ETFB Entrez, Source | electron-transfer-flavoprotein, beta polypeptide | 865 | 0.212 | 0.4772 | Yes |
| 12 | ATP5G1 | ATP5G1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) | 878 | 0.210 | 0.5009 | Yes |
| 13 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 880 | 0.210 | 0.5254 | Yes |
| 14 | DBI | DBI Entrez, Source | diazepam binding inhibitor (GABA receptor modulator, acyl-Coenzyme A binding protein) | 1077 | 0.184 | 0.5323 | Yes |
| 15 | DNAJB9 | DNAJB9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 9 | 1133 | 0.178 | 0.5491 | Yes |
| 16 | CHP | CHP Entrez, Source | - | 1193 | 0.171 | 0.5647 | Yes |
| 17 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 1274 | 0.166 | 0.5781 | Yes |
| 18 | RASD1 | RASD1 Entrez, Source | RAS, dexamethasone-induced 1 | 1396 | 0.157 | 0.5874 | Yes |
| 19 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.6053 | Yes |
| 20 | ARL6IP2 | ARL6IP2 Entrez, Source | ADP-ribosylation factor-like 6 interacting protein 2 | 1624 | 0.142 | 0.6054 | Yes |
| 21 | COX17 | COX17 Entrez, Source | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) | 1921 | 0.125 | 0.5978 | Yes |
| 22 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 1940 | 0.124 | 0.6110 | Yes |
| 23 | GRPEL1 | GRPEL1 Entrez, Source | GrpE-like 1, mitochondrial (E. coli) | 2017 | 0.121 | 0.6195 | Yes |
| 24 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 2248 | 0.113 | 0.6154 | Yes |
| 25 | TALDO1 | TALDO1 Entrez, Source | transaldolase 1 | 2443 | 0.105 | 0.6131 | Yes |
| 26 | GPHN | GPHN Entrez, Source | gephyrin | 2447 | 0.105 | 0.6251 | Yes |
| 27 | PPARG | PPARG Entrez, Source | peroxisome proliferative activated receptor, gamma | 2538 | 0.101 | 0.6302 | Yes |
| 28 | PEX14 | PEX14 Entrez, Source | peroxisomal biogenesis factor 14 | 2722 | 0.095 | 0.6276 | Yes |
| 29 | IFNGR1 | IFNGR1 Entrez, Source | interferon gamma receptor 1 | 2743 | 0.094 | 0.6371 | Yes |
| 30 | MKNK2 | MKNK2 Entrez, Source | MAP kinase interacting serine/threonine kinase 2 | 2879 | 0.089 | 0.6373 | Yes |
| 31 | PDCD8 | PDCD8 Entrez, Source | programmed cell death 8 (apoptosis-inducing factor) | 3003 | 0.085 | 0.6380 | Yes |
| 32 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 3857 | 0.063 | 0.5812 | No |
| 33 | PIM3 | PIM3 Entrez, Source | pim-3 oncogene | 3946 | 0.061 | 0.5817 | No |
| 34 | PHB | PHB Entrez, Source | prohibitin | 3983 | 0.060 | 0.5860 | No |
| 35 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4310 | 0.053 | 0.5676 | No |
| 36 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.5705 | No |
| 37 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 4456 | 0.049 | 0.5686 | No |
| 38 | PDIA6 | PDIA6 Entrez, Source | protein disulfide isomerase family A, member 6 | 5150 | 0.035 | 0.5205 | No |
| 39 | MRPL34 | MRPL34 Entrez, Source | mitochondrial ribosomal protein L34 | 5252 | 0.033 | 0.5167 | No |
| 40 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 5716 | 0.023 | 0.4846 | No |
| 41 | COX4NB | COX4NB Entrez, Source | COX4 neighbor | 5788 | 0.022 | 0.4819 | No |
| 42 | SLC5A6 | SLC5A6 Entrez, Source | solute carrier family 5 (sodium-dependent vitamin transporter), member 6 | 5944 | 0.019 | 0.4725 | No |
| 43 | USMG5 | USMG5 Entrez, Source | upregulated during skeletal muscle growth 5 homolog (mouse) | 6069 | 0.017 | 0.4651 | No |
| 44 | PHB2 | PHB2 Entrez, Source | prohibitin 2 | 6088 | 0.016 | 0.4657 | No |
| 45 | PSMA1 | PSMA1 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 1 | 6413 | 0.009 | 0.4424 | No |
| 46 | ESRRA | ESRRA Entrez, Source | estrogen-related receptor alpha | 6502 | 0.008 | 0.4367 | No |
| 47 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 6598 | 0.006 | 0.4302 | No |
| 48 | COX6A1 | COX6A1 Entrez, Source | cytochrome c oxidase subunit VIa polypeptide 1 | 6867 | 0.001 | 0.4102 | No |
| 49 | MAPK6 | MAPK6 Entrez, Source | mitogen-activated protein kinase 6 | 7114 | -0.004 | 0.3922 | No |
| 50 | NDUFA8 | NDUFA8 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa | 7295 | -0.008 | 0.3796 | No |
| 51 | PSMA5 | PSMA5 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 5 | 7365 | -0.010 | 0.3755 | No |
| 52 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 7996 | -0.023 | 0.3308 | No |
| 53 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 8212 | -0.028 | 0.3179 | No |
| 54 | LRRC59 | LRRC59 Entrez, Source | leucine rich repeat containing 59 | 8742 | -0.040 | 0.2827 | No |
| 55 | BCAP31 | BCAP31 Entrez, Source | B-cell receptor-associated protein 31 | 9224 | -0.052 | 0.2526 | No |
| 56 | XBP1 | XBP1 Entrez, Source | X-box binding protein 1 | 9264 | -0.053 | 0.2559 | No |
| 57 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9745 | -0.066 | 0.2275 | No |
| 58 | TXNDC14 | TXNDC14 Entrez, Source | thioredoxin domain containing 14 | 10234 | -0.082 | 0.2005 | No |
| 59 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 10317 | -0.085 | 0.2043 | No |
| 60 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 10416 | -0.089 | 0.2073 | No |
| 61 | FDX1 | FDX1 Entrez, Source | ferredoxin 1 | 10969 | -0.110 | 0.1787 | No |