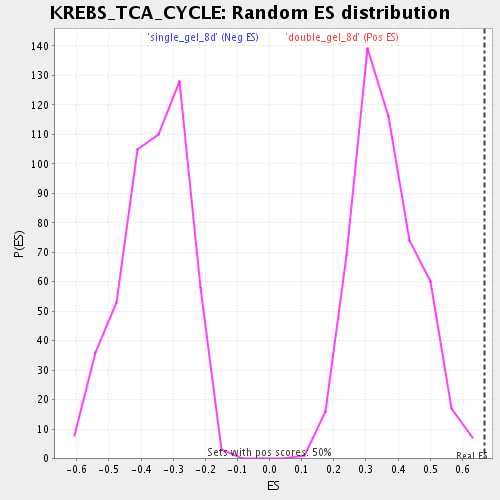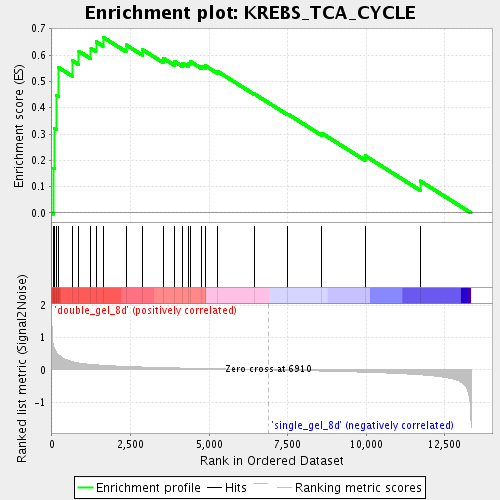
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | KREBS_TCA_CYCLE |
| Enrichment Score (ES) | 0.6677192 |
| Normalized Enrichment Score (NES) | 1.8364694 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009777463 |
| FWER p-Value | 0.762 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 58 | 0.726 | 0.1686 | Yes |
| 2 | PC | PC Entrez, Source | pyruvate carboxylase | 79 | 0.652 | 0.3226 | Yes |
| 3 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 143 | 0.545 | 0.4479 | Yes |
| 4 | PDP2 | PDP2 Entrez, Source | - | 217 | 0.460 | 0.5521 | Yes |
| 5 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 663 | 0.250 | 0.5782 | Yes |
| 6 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.6143 | Yes |
| 7 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 1243 | 0.168 | 0.6254 | Yes |
| 8 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.6508 | Yes |
| 9 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 1628 | 0.141 | 0.6677 | Yes |
| 10 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2361 | 0.108 | 0.6386 | No |
| 11 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.6203 | No |
| 12 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 3545 | 0.070 | 0.5875 | No |
| 13 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.5759 | No |
| 14 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 4168 | 0.056 | 0.5690 | No |
| 15 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.5675 | No |
| 16 | SUCLG2 | SUCLG2 Entrez, Source | succinate-CoA ligase, GDP-forming, beta subunit | 4416 | 0.050 | 0.5748 | No |
| 17 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 4772 | 0.042 | 0.5582 | No |
| 18 | DLST | DLST Entrez, Source | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 4884 | 0.040 | 0.5595 | No |
| 19 | SUCLG1 | SUCLG1 Entrez, Source | succinate-CoA ligase, GDP-forming, alpha subunit | 5260 | 0.033 | 0.5391 | No |
| 20 | IDH3A | IDH3A Entrez, Source | isocitrate dehydrogenase 3 (NAD+) alpha | 6433 | 0.009 | 0.4533 | No |
| 21 | CS | CS Entrez, Source | citrate synthase | 7513 | -0.013 | 0.3753 | No |
| 22 | IDH3G | IDH3G Entrez, Source | isocitrate dehydrogenase 3 (NAD+) gamma | 8587 | -0.037 | 0.3034 | No |
| 23 | PPM2C | PPM2C Entrez, Source | protein phosphatase 2C, magnesium-dependent, catalytic subunit | 9971 | -0.074 | 0.2172 | No |
| 24 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 11723 | -0.150 | 0.1216 | No |