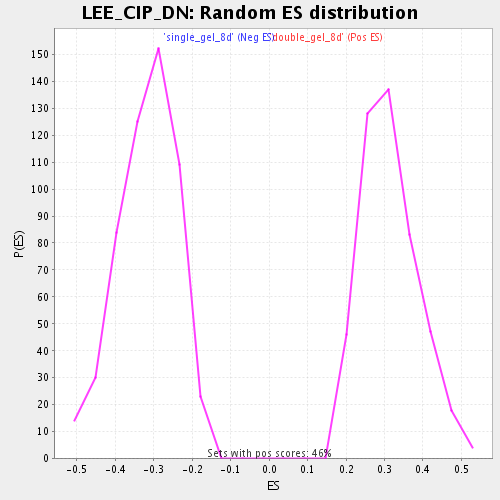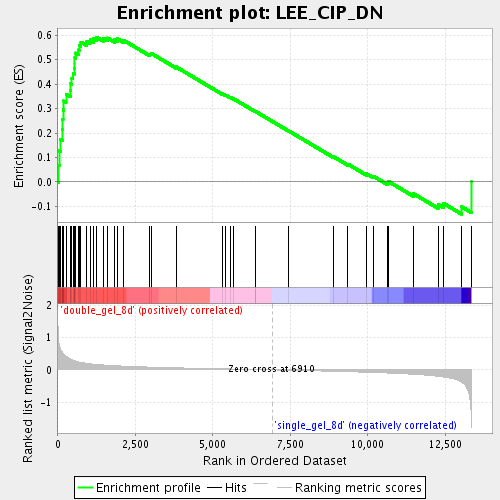
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | LEE_CIP_DN |
| Enrichment Score (ES) | 0.59125775 |
| Normalized Enrichment Score (NES) | 1.8817227 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0063762846 |
| FWER p-Value | 0.541 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 21 | 0.893 | 0.0688 | Yes |
| 2 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 49 | 0.765 | 0.1272 | Yes |
| 3 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 87 | 0.637 | 0.1746 | Yes |
| 4 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 140 | 0.550 | 0.2141 | Yes |
| 5 | PPP1R3C | PPP1R3C Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 3C | 146 | 0.543 | 0.2565 | Yes |
| 6 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 167 | 0.505 | 0.2949 | Yes |
| 7 | PON1 | PON1 Entrez, Source | paraoxonase 1 | 193 | 0.481 | 0.3310 | Yes |
| 8 | MBL2 | MBL2 Entrez, Source | mannose-binding lectin (protein C) 2, soluble (opsonic defect) | 263 | 0.424 | 0.3593 | Yes |
| 9 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 411 | 0.336 | 0.3747 | Yes |
| 10 | C4BPA | C4BPA Entrez, Source | complement component 4 binding protein, alpha | 417 | 0.334 | 0.4007 | Yes |
| 11 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 450 | 0.320 | 0.4235 | Yes |
| 12 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 487 | 0.306 | 0.4449 | Yes |
| 13 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 540 | 0.287 | 0.4637 | Yes |
| 14 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 548 | 0.283 | 0.4854 | Yes |
| 15 | HAL | HAL Entrez, Source | histidine ammonia-lyase | 549 | 0.282 | 0.5077 | Yes |
| 16 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 580 | 0.273 | 0.5270 | Yes |
| 17 | SLC37A4 | SLC37A4 Entrez, Source | solute carrier family 37 (glycerol-6-phosphate transporter), member 4 | 675 | 0.246 | 0.5393 | Yes |
| 18 | SLC17A3 | SLC17A3 Entrez, Source | solute carrier family 17 (sodium phosphate), member 3 | 687 | 0.241 | 0.5575 | Yes |
| 19 | SULT1B1 | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 743 | 0.229 | 0.5715 | Yes |
| 20 | CYP7A1 | CYP7A1 Entrez, Source | cytochrome P450, family 7, subfamily A, polypeptide 1 | 934 | 0.202 | 0.5731 | Yes |
| 21 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 1037 | 0.188 | 0.5803 | Yes |
| 22 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1158 | 0.175 | 0.5851 | Yes |
| 23 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 1252 | 0.167 | 0.5913 | Yes |
| 24 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 1460 | 0.152 | 0.5877 | No |
| 25 | TRPV2 | TRPV2 Entrez, Source | transient receptor potential cation channel, subfamily V, member 2 | 1604 | 0.143 | 0.5882 | No |
| 26 | SLC1A2 | SLC1A2 Entrez, Source | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 1833 | 0.129 | 0.5813 | No |
| 27 | DPP4 | DPP4 Entrez, Source | dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) | 1919 | 0.125 | 0.5848 | No |
| 28 | TNNT3 | TNNT3 Entrez, Source | troponin T type 3 (skeletal, fast) | 2118 | 0.117 | 0.5791 | No |
| 29 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 2969 | 0.086 | 0.5219 | No |
| 30 | TNNI2 | TNNI2 Entrez, Source | troponin I type 2 (skeletal, fast) | 3007 | 0.085 | 0.5258 | No |
| 31 | DMBT1 | DMBT1 Entrez, Source | deleted in malignant brain tumors 1 | 3812 | 0.064 | 0.4704 | No |
| 32 | CMAH | CMAH Entrez, Source | cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) | 5295 | 0.032 | 0.3614 | No |
| 33 | PNLIPRP1 | PNLIPRP1 Entrez, Source | pancreatic lipase-related protein 1 | 5421 | 0.030 | 0.3544 | No |
| 34 | PRODH | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 5569 | 0.026 | 0.3454 | No |
| 35 | CYP26A1 | CYP26A1 Entrez, Source | cytochrome P450, family 26, subfamily A, polypeptide 1 | 5664 | 0.024 | 0.3402 | No |
| 36 | CTRC | CTRC Entrez, Source | chymotrypsin C (caldecrin) | 6373 | 0.010 | 0.2878 | No |
| 37 | ELA2 | ELA2 Entrez, Source | elastase 2, neutrophil | 6385 | 0.010 | 0.2877 | No |
| 38 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 7432 | -0.011 | 0.2099 | No |
| 39 | MAPKAPK2 | MAPKAPK2 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 2 | 8907 | -0.044 | 0.1025 | No |
| 40 | PNLIP | PNLIP Entrez, Source | pancreatic lipase | 9358 | -0.056 | 0.0730 | No |
| 41 | GSTA4 | GSTA4 Entrez, Source | glutathione S-transferase A4 | 9961 | -0.073 | 0.0336 | No |
| 42 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 10171 | -0.080 | 0.0242 | No |
| 43 | SELENBP1 | SELENBP1 Entrez, Source | selenium binding protein 1 | 10631 | -0.096 | -0.0028 | No |
| 44 | HBP1 | HBP1 Entrez, Source | HMG-box transcription factor 1 | 10662 | -0.097 | 0.0027 | No |
| 45 | GPR37 | GPR37 Entrez, Source | G protein-coupled receptor 37 (endothelin receptor type B-like) | 11472 | -0.135 | -0.0475 | No |
| 46 | RNASE4 | RNASE4 Entrez, Source | ribonuclease, RNase A family, 4 | 12272 | -0.201 | -0.0917 | No |
| 47 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 12440 | -0.220 | -0.0870 | No |
| 48 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13028 | -0.381 | -0.1011 | No |
| 49 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13339 | -1.580 | 0.0002 | No |