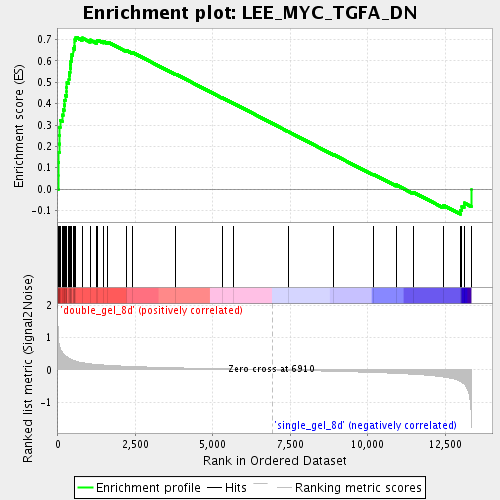
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | LEE_MYC_TGFA_DN |
| Enrichment Score (ES) | 0.7112373 |
| Normalized Enrichment Score (NES) | 2.3386533 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | G0S2 | G0S2 Entrez, Source | G0/G1switch 2 | 6 | 1.218 | 0.0632 | Yes |
| 2 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 8 | 1.205 | 0.1261 | Yes |
| 3 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 21 | 0.893 | 0.1718 | Yes |
| 4 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 42 | 0.784 | 0.2112 | Yes |
| 5 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 49 | 0.765 | 0.2508 | Yes |
| 6 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 56 | 0.731 | 0.2885 | Yes |
| 7 | FABP2 | FABP2 Entrez, Source | fatty acid binding protein 2, intestinal | 71 | 0.690 | 0.3235 | Yes |
| 8 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 140 | 0.550 | 0.3471 | Yes |
| 9 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 167 | 0.505 | 0.3715 | Yes |
| 10 | KLF15 | KLF15 Entrez, Source | Kruppel-like factor 15 | 200 | 0.476 | 0.3940 | Yes |
| 11 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 216 | 0.460 | 0.4169 | Yes |
| 12 | IGF1 | IGF1 Entrez, Source | insulin-like growth factor 1 (somatomedin C) | 236 | 0.445 | 0.4387 | Yes |
| 13 | HLA-DMA | HLA-DMA Entrez, Source | major histocompatibility complex, class II, DM alpha | 281 | 0.413 | 0.4570 | Yes |
| 14 | KHK | KHK Entrez, Source | ketohexokinase (fructokinase) | 285 | 0.407 | 0.4780 | Yes |
| 15 | CPT2 | CPT2 Entrez, Source | carnitine palmitoyltransferase II | 292 | 0.402 | 0.4986 | Yes |
| 16 | TDO2 | TDO2 Entrez, Source | tryptophan 2,3-dioxygenase | 355 | 0.360 | 0.5127 | Yes |
| 17 | HADH2 | HADH2 Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase, type II | 361 | 0.357 | 0.5310 | Yes |
| 18 | PXMP2 | PXMP2 Entrez, Source | peroxisomal membrane protein 2, 22kDa | 380 | 0.350 | 0.5480 | Yes |
| 19 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 392 | 0.341 | 0.5650 | Yes |
| 20 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 403 | 0.337 | 0.5818 | Yes |
| 21 | C4BPA | C4BPA Entrez, Source | complement component 4 binding protein, alpha | 417 | 0.334 | 0.5983 | Yes |
| 22 | ACOX1 | ACOX1 Entrez, Source | acyl-Coenzyme A oxidase 1, palmitoyl | 429 | 0.329 | 0.6147 | Yes |
| 23 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 450 | 0.320 | 0.6299 | Yes |
| 24 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 487 | 0.306 | 0.6432 | Yes |
| 25 | SLC25A22 | SLC25A22 Entrez, Source | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 488 | 0.305 | 0.6591 | Yes |
| 26 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 540 | 0.287 | 0.6702 | Yes |
| 27 | KYNU | KYNU Entrez, Source | kynureninase (L-kynurenine hydrolase) | 548 | 0.283 | 0.6845 | Yes |
| 28 | HAL | HAL Entrez, Source | histidine ammonia-lyase | 549 | 0.282 | 0.6992 | Yes |
| 29 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 580 | 0.273 | 0.7112 | Yes |
| 30 | IGFALS | IGFALS Entrez, Source | insulin-like growth factor binding protein, acid labile subunit | 780 | 0.223 | 0.7079 | No |
| 31 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 1037 | 0.188 | 0.6985 | No |
| 32 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 1252 | 0.167 | 0.6911 | No |
| 33 | AQP8 | AQP8 Entrez, Source | aquaporin 8 | 1275 | 0.166 | 0.6981 | No |
| 34 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 1460 | 0.152 | 0.6922 | No |
| 35 | TRPV2 | TRPV2 Entrez, Source | transient receptor potential cation channel, subfamily V, member 2 | 1604 | 0.143 | 0.6889 | No |
| 36 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 2203 | 0.114 | 0.6499 | No |
| 37 | ELOVL5 | ELOVL5 Entrez, Source | ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 2393 | 0.107 | 0.6413 | No |
| 38 | ABCG2 | ABCG2 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 2 | 3807 | 0.064 | 0.5383 | No |
| 39 | CMAH | CMAH Entrez, Source | cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) | 5295 | 0.032 | 0.4281 | No |
| 40 | PSEN2 | PSEN2 Entrez, Source | presenilin 2 (Alzheimer disease 4) | 5670 | 0.024 | 0.4013 | No |
| 41 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 7432 | -0.011 | 0.2693 | No |
| 42 | MAPKAPK2 | MAPKAPK2 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 2 | 8907 | -0.044 | 0.1607 | No |
| 43 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 10171 | -0.080 | 0.0699 | No |
| 44 | CRISP2 | CRISP2 Entrez, Source | cysteine-rich secretory protein 2 | 10913 | -0.108 | 0.0198 | No |
| 45 | GPR37 | GPR37 Entrez, Source | G protein-coupled receptor 37 (endothelin receptor type B-like) | 11472 | -0.135 | -0.0151 | No |
| 46 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 12440 | -0.220 | -0.0764 | No |
| 47 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 12985 | -0.357 | -0.0986 | No |
| 48 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 13038 | -0.386 | -0.0824 | No |
| 49 | LIPC | LIPC Entrez, Source | lipase, hepatic | 13109 | -0.433 | -0.0651 | No |
| 50 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13339 | -1.580 | 0.0002 | No |