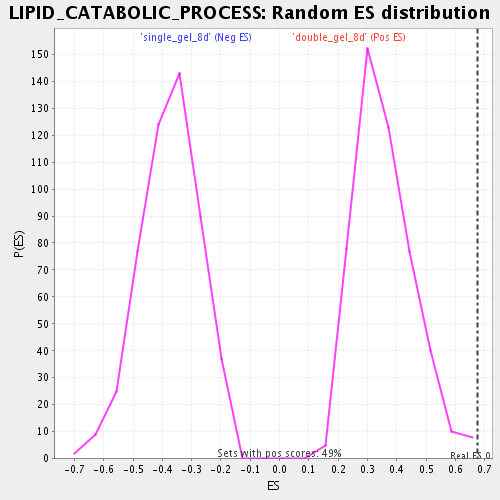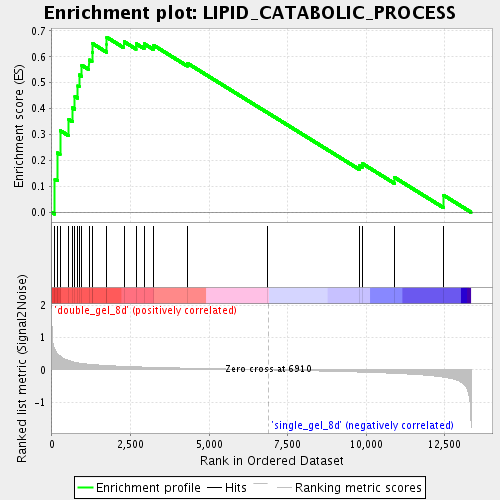
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | LIPID_CATABOLIC_PROCESS |
| Enrichment Score (ES) | 0.67580265 |
| Normalized Enrichment Score (NES) | 1.8870798 |
| Nominal p-value | 0.0020283975 |
| FDR q-value | 0.0062071234 |
| FWER p-Value | 0.516 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | APOA4 | APOA4 Entrez, Source | apolipoprotein A-IV | 96 | 0.620 | 0.1274 | Yes |
| 2 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 177 | 0.496 | 0.2291 | Yes |
| 3 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.3157 | Yes |
| 4 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 537 | 0.288 | 0.3575 | Yes |
| 5 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 656 | 0.251 | 0.4032 | Yes |
| 6 | PPARA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 732 | 0.231 | 0.4477 | Yes |
| 7 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 827 | 0.217 | 0.4876 | Yes |
| 8 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 870 | 0.211 | 0.5304 | Yes |
| 9 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 950 | 0.200 | 0.5679 | Yes |
| 10 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 1180 | 0.172 | 0.5881 | Yes |
| 11 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 1278 | 0.165 | 0.6167 | Yes |
| 12 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1296 | 0.164 | 0.6512 | Yes |
| 13 | STS | STS Entrez, Source | steroid sulfatase (microsomal), arylsulfatase C, isozyme S | 1743 | 0.134 | 0.6469 | Yes |
| 14 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1747 | 0.134 | 0.6758 | Yes |
| 15 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 2294 | 0.111 | 0.6588 | No |
| 16 | CEL | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 2678 | 0.096 | 0.6510 | No |
| 17 | NEU3 | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 2937 | 0.087 | 0.6505 | No |
| 18 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 3231 | 0.078 | 0.6453 | No |
| 19 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4310 | 0.053 | 0.5758 | No |
| 20 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 6850 | 0.001 | 0.3855 | No |
| 21 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 9794 | -0.068 | 0.1792 | No |
| 22 | PPT1 | PPT1 Entrez, Source | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 9878 | -0.071 | 0.1883 | No |
| 23 | LYPLA3 | LYPLA3 Entrez, Source | lysophospholipase 3 (lysosomal phospholipase A2) | 10907 | -0.107 | 0.1344 | No |
| 24 | SMPD3 | SMPD3 Entrez, Source | sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | 12464 | -0.223 | 0.0659 | No |