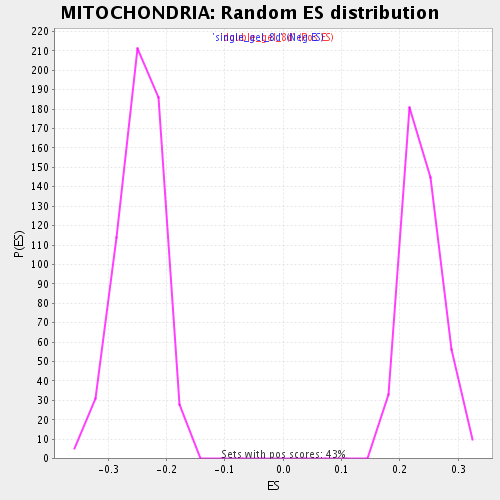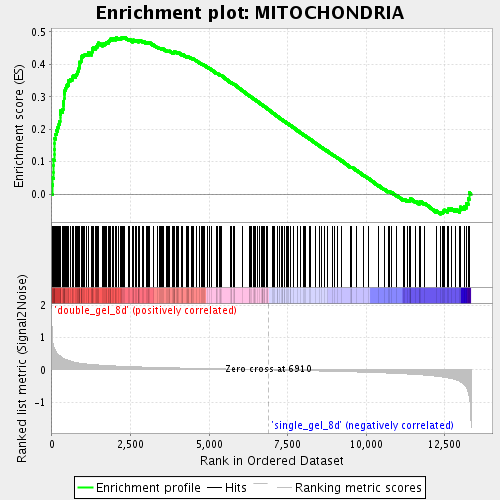
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | MITOCHONDRIA |
| Enrichment Score (ES) | 0.4835339 |
| Normalized Enrichment Score (NES) | 2.0315182 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0012169083 |
| FWER p-Value | 0.089 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 10 | 1.121 | 0.0284 | Yes |
| 2 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 24 | 0.864 | 0.0499 | Yes |
| 3 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 49 | 0.765 | 0.0680 | Yes |
| 4 | BCL2A1 | BCL2A1 Entrez, Source | BCL2-related protein A1 | 53 | 0.740 | 0.0870 | Yes |
| 5 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 58 | 0.726 | 0.1056 | Yes |
| 6 | LCAT | LCAT Entrez, Source | lecithin-cholesterol acyltransferase | 76 | 0.677 | 0.1219 | Yes |
| 7 | PC | PC Entrez, Source | pyruvate carboxylase | 79 | 0.652 | 0.1387 | Yes |
| 8 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 87 | 0.637 | 0.1548 | Yes |
| 9 | CPS1 | CPS1 Entrez, Source | carbamoyl-phosphate synthetase 1, mitochondrial | 91 | 0.631 | 0.1709 | Yes |
| 10 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 113 | 0.597 | 0.1849 | Yes |
| 11 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 143 | 0.545 | 0.1968 | Yes |
| 12 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 184 | 0.487 | 0.2065 | Yes |
| 13 | TAT | TAT Entrez, Source | tyrosine aminotransferase | 221 | 0.457 | 0.2156 | Yes |
| 14 | SARDH | SARDH Entrez, Source | sarcosine dehydrogenase | 245 | 0.438 | 0.2252 | Yes |
| 15 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 259 | 0.427 | 0.2353 | Yes |
| 16 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.2465 | Yes |
| 17 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 271 | 0.419 | 0.2566 | Yes |
| 18 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 344 | 0.371 | 0.2607 | Yes |
| 19 | HMGCS2 | HMGCS2 Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | 360 | 0.358 | 0.2689 | Yes |
| 20 | ETFDH | ETFDH Entrez, Source | electron-transferring-flavoprotein dehydrogenase | 363 | 0.357 | 0.2780 | Yes |
| 21 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 374 | 0.353 | 0.2865 | Yes |
| 22 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 384 | 0.346 | 0.2948 | Yes |
| 23 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 389 | 0.344 | 0.3034 | Yes |
| 24 | SOD2 | SOD2 Entrez, Source | superoxide dismutase 2, mitochondrial | 391 | 0.343 | 0.3123 | Yes |
| 25 | TIMM8A | TIMM8A Entrez, Source | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 412 | 0.336 | 0.3195 | Yes |
| 26 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 430 | 0.329 | 0.3268 | Yes |
| 27 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 450 | 0.320 | 0.3336 | Yes |
| 28 | SLC25A22 | SLC25A22 Entrez, Source | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 488 | 0.305 | 0.3387 | Yes |
| 29 | AMACR | AMACR Entrez, Source | alpha-methylacyl-CoA racemase | 533 | 0.289 | 0.3429 | Yes |
| 30 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 537 | 0.288 | 0.3502 | Yes |
| 31 | CYP27A1 | CYP27A1 Entrez, Source | cytochrome P450, family 27, subfamily A, polypeptide 1 | 578 | 0.274 | 0.3542 | Yes |
| 32 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 642 | 0.255 | 0.3561 | Yes |
| 33 | DCI | DCI Entrez, Source | dodecenoyl-Coenzyme A delta isomerase (3,2 trans-enoyl-Coenzyme A isomerase) | 660 | 0.250 | 0.3613 | Yes |
| 34 | FXC1 | FXC1 Entrez, Source | fracture callus 1 homolog (rat) | 696 | 0.239 | 0.3648 | Yes |
| 35 | SLC25A20 | SLC25A20 Entrez, Source | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 | 759 | 0.226 | 0.3660 | Yes |
| 36 | CLPX | CLPX Entrez, Source | ClpX caseinolytic peptidase X homolog (E. coli) | 785 | 0.221 | 0.3698 | Yes |
| 37 | DBT | DBT Entrez, Source | dihydrolipoamide branched chain transacylase E2 | 799 | 0.220 | 0.3746 | Yes |
| 38 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 828 | 0.216 | 0.3781 | Yes |
| 39 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 839 | 0.215 | 0.3829 | Yes |
| 40 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.3870 | Yes |
| 41 | ETFB | ETFB Entrez, Source | electron-transfer-flavoprotein, beta polypeptide | 865 | 0.212 | 0.3921 | Yes |
| 42 | ATP5G1 | ATP5G1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) | 878 | 0.210 | 0.3967 | Yes |
| 43 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 880 | 0.210 | 0.4020 | Yes |
| 44 | CAT | CAT Entrez, Source | catalase | 884 | 0.209 | 0.4073 | Yes |
| 45 | TST | TST Entrez, Source | thiosulfate sulfurtransferase (rhodanese) | 926 | 0.203 | 0.4094 | Yes |
| 46 | CYP7A1 | CYP7A1 Entrez, Source | cytochrome P450, family 7, subfamily A, polypeptide 1 | 934 | 0.202 | 0.4141 | Yes |
| 47 | SLC25A1 | SLC25A1 Entrez, Source | solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 | 946 | 0.200 | 0.4185 | Yes |
| 48 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 950 | 0.200 | 0.4235 | Yes |
| 49 | GNAS | GNAS Entrez, Source | GNAS complex locus | 968 | 0.198 | 0.4273 | Yes |
| 50 | TXN2 | TXN2 Entrez, Source | thioredoxin 2 | 1019 | 0.190 | 0.4285 | Yes |
| 51 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 1050 | 0.187 | 0.4310 | Yes |
| 52 | SHMT2 | SHMT2 Entrez, Source | serine hydroxymethyltransferase 2 (mitochondrial) | 1105 | 0.181 | 0.4316 | Yes |
| 53 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1158 | 0.175 | 0.4322 | Yes |
| 54 | NDUFV2 | NDUFV2 Entrez, Source | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | 1161 | 0.175 | 0.4366 | Yes |
| 55 | CABC1 | CABC1 Entrez, Source | chaperone, ABC1 activity of bc1 complex homolog (S. pombe) | 1268 | 0.166 | 0.4328 | Yes |
| 56 | PRDX3 | PRDX3 Entrez, Source | peroxiredoxin 3 | 1274 | 0.166 | 0.4367 | Yes |
| 57 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 1278 | 0.165 | 0.4408 | Yes |
| 58 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 1288 | 0.165 | 0.4444 | Yes |
| 59 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1296 | 0.164 | 0.4482 | Yes |
| 60 | DECR1 | DECR1 Entrez, Source | 2,4-dienoyl CoA reductase 1, mitochondrial | 1315 | 0.163 | 0.4510 | Yes |
| 61 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1399 | 0.157 | 0.4488 | Yes |
| 62 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.4525 | Yes |
| 63 | TFB2M | TFB2M Entrez, Source | transcription factor B2, mitochondrial | 1418 | 0.156 | 0.4556 | Yes |
| 64 | AFG3L2 | AFG3L2 Entrez, Source | AFG3 ATPase family gene 3-like 2 (yeast) | 1449 | 0.153 | 0.4573 | Yes |
| 65 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 1460 | 0.152 | 0.4605 | Yes |
| 66 | COQ7 | COQ7 Entrez, Source | coenzyme Q7 homolog, ubiquinone (yeast) | 1476 | 0.151 | 0.4633 | Yes |
| 67 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 1491 | 0.150 | 0.4661 | Yes |
| 68 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 1620 | 0.142 | 0.4600 | Yes |
| 69 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 1628 | 0.141 | 0.4632 | Yes |
| 70 | PECI | PECI Entrez, Source | peroxisomal D3,D2-enoyl-CoA isomerase | 1680 | 0.138 | 0.4629 | Yes |
| 71 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1712 | 0.137 | 0.4640 | Yes |
| 72 | PLA2G2A | PLA2G2A Entrez, Source | phospholipase A2, group IIA (platelets, synovial fluid) | 1721 | 0.136 | 0.4670 | Yes |
| 73 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1747 | 0.134 | 0.4685 | Yes |
| 74 | SPG7 | SPG7 Entrez, Source | spastic paraplegia 7, paraplegin (pure and complicated autosomal recessive) | 1792 | 0.132 | 0.4686 | Yes |
| 75 | SLC25A21 | SLC25A21 Entrez, Source | solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 | 1804 | 0.131 | 0.4712 | Yes |
| 76 | SLC1A2 | SLC1A2 Entrez, Source | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 1833 | 0.129 | 0.4724 | Yes |
| 77 | COX7B | COX7B Entrez, Source | cytochrome c oxidase subunit VIIb | 1849 | 0.129 | 0.4746 | Yes |
| 78 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 1864 | 0.128 | 0.4769 | Yes |
| 79 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 1880 | 0.127 | 0.4790 | Yes |
| 80 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 1940 | 0.124 | 0.4778 | Yes |
| 81 | MIPEP | MIPEP Entrez, Source | mitochondrial intermediate peptidase | 1964 | 0.123 | 0.4792 | Yes |
| 82 | ACADL | ACADL Entrez, Source | acyl-Coenzyme A dehydrogenase, long chain | 2024 | 0.121 | 0.4778 | Yes |
| 83 | ENDOG | ENDOG Entrez, Source | endonuclease G | 2036 | 0.120 | 0.4801 | Yes |
| 84 | MRPL46 | MRPL46 Entrez, Source | mitochondrial ribosomal protein L46 | 2040 | 0.120 | 0.4830 | Yes |
| 85 | MRPL16 | MRPL16 Entrez, Source | mitochondrial ribosomal protein L16 | 2132 | 0.117 | 0.4791 | Yes |
| 86 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2183 | 0.115 | 0.4783 | Yes |
| 87 | TXNRD2 | TXNRD2 Entrez, Source | thioredoxin reductase 2 | 2200 | 0.114 | 0.4800 | Yes |
| 88 | MB | MB Entrez, Source | myoglobin | 2219 | 0.114 | 0.4816 | Yes |
| 89 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 2248 | 0.113 | 0.4824 | Yes |
| 90 | BCS1L | BCS1L Entrez, Source | BCS1-like (yeast) | 2272 | 0.111 | 0.4835 | Yes |
| 91 | ATP5E | ATP5E Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | 2324 | 0.109 | 0.4825 | No |
| 92 | ATP5O | ATP5O Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) | 2450 | 0.105 | 0.4757 | No |
| 93 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 2467 | 0.104 | 0.4771 | No |
| 94 | TFB1M | TFB1M Entrez, Source | transcription factor B1, mitochondrial | 2580 | 0.100 | 0.4712 | No |
| 95 | TIMM8B | TIMM8B Entrez, Source | translocase of inner mitochondrial membrane 8 homolog B (yeast) | 2592 | 0.100 | 0.4729 | No |
| 96 | POLR1B | POLR1B Entrez, Source | polymerase (RNA) I polypeptide B, 128kDa | 2596 | 0.099 | 0.4753 | No |
| 97 | ETFA | ETFA Entrez, Source | electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) | 2650 | 0.098 | 0.4738 | No |
| 98 | STAR | STAR Entrez, Source | steroidogenic acute regulator | 2676 | 0.096 | 0.4744 | No |
| 99 | DHODH | DHODH Entrez, Source | dihydroorotate dehydrogenase | 2752 | 0.094 | 0.4711 | No |
| 100 | SLC25A5 | SLC25A5 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 2756 | 0.094 | 0.4733 | No |
| 101 | GPD2 | GPD2 Entrez, Source | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) | 2771 | 0.093 | 0.4746 | No |
| 102 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 2802 | 0.091 | 0.4747 | No |
| 103 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.4707 | No |
| 104 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 2926 | 0.087 | 0.4700 | No |
| 105 | PDCD8 | PDCD8 Entrez, Source | programmed cell death 8 (apoptosis-inducing factor) | 3003 | 0.085 | 0.4664 | No |
| 106 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 3038 | 0.084 | 0.4659 | No |
| 107 | TOMM70A | TOMM70A Entrez, Source | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | 3062 | 0.083 | 0.4663 | No |
| 108 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 3082 | 0.082 | 0.4670 | No |
| 109 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 3098 | 0.082 | 0.4680 | No |
| 110 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 3227 | 0.078 | 0.4602 | No |
| 111 | MRPL41 | MRPL41 Entrez, Source | mitochondrial ribosomal protein L41 | 3347 | 0.075 | 0.4531 | No |
| 112 | SNCA | SNCA Entrez, Source | synuclein, alpha (non A4 component of amyloid precursor) | 3411 | 0.073 | 0.4502 | No |
| 113 | ARG2 | ARG2 Entrez, Source | arginase, type II | 3449 | 0.072 | 0.4492 | No |
| 114 | MRPS15 | MRPS15 Entrez, Source | mitochondrial ribosomal protein S15 | 3496 | 0.071 | 0.4476 | No |
| 115 | COX6A2 | COX6A2 Entrez, Source | cytochrome c oxidase subunit VIa polypeptide 2 | 3517 | 0.071 | 0.4479 | No |
| 116 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 3545 | 0.070 | 0.4476 | No |
| 117 | COX7A2 | COX7A2 Entrez, Source | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | 3647 | 0.068 | 0.4417 | No |
| 118 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 3691 | 0.067 | 0.4401 | No |
| 119 | ATP5J | ATP5J Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 3709 | 0.066 | 0.4406 | No |
| 120 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 3731 | 0.066 | 0.4407 | No |
| 121 | MRPS7 | MRPS7 Entrez, Source | mitochondrial ribosomal protein S7 | 3745 | 0.065 | 0.4414 | No |
| 122 | RAF1 | RAF1 Entrez, Source | v-raf-1 murine leukemia viral oncogene homolog 1 | 3847 | 0.063 | 0.4353 | No |
| 123 | AMT | AMT Entrez, Source | aminomethyltransferase | 3875 | 0.063 | 0.4349 | No |
| 124 | VAV1 | VAV1 Entrez, Source | vav 1 oncogene | 3880 | 0.062 | 0.4362 | No |
| 125 | ATP5F1 | ATP5F1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 3886 | 0.062 | 0.4374 | No |
| 126 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3898 | 0.062 | 0.4382 | No |
| 127 | NDUFC2 | NDUFC2 Entrez, Source | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa | 3909 | 0.062 | 0.4390 | No |
| 128 | ATP5I | ATP5I Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit E | 3963 | 0.061 | 0.4366 | No |
| 129 | PRSS15 | PRSS15 Entrez, Source | protease, serine, 15 | 3990 | 0.060 | 0.4361 | No |
| 130 | BNIP3L | BNIP3L Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3-like | 4011 | 0.059 | 0.4361 | No |
| 131 | UQCRFS1 | UQCRFS1 Entrez, Source | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 4024 | 0.059 | 0.4367 | No |
| 132 | UCP1 | UCP1 Entrez, Source | uncoupling protein 1 (mitochondrial, proton carrier) | 4133 | 0.056 | 0.4300 | No |
| 133 | UQCRC2 | UQCRC2 Entrez, Source | ubiquinol-cytochrome c reductase core protein II | 4141 | 0.056 | 0.4309 | No |
| 134 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 4168 | 0.056 | 0.4303 | No |
| 135 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 4283 | 0.053 | 0.4230 | No |
| 136 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4310 | 0.053 | 0.4224 | No |
| 137 | COX5B | COX5B Entrez, Source | cytochrome c oxidase subunit Vb | 4325 | 0.052 | 0.4227 | No |
| 138 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | 0.4220 | No |
| 139 | NDUFS6 | NDUFS6 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) | 4355 | 0.051 | 0.4232 | No |
| 140 | COX6C | COX6C Entrez, Source | cytochrome c oxidase subunit VIc | 4441 | 0.050 | 0.4180 | No |
| 141 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 4456 | 0.049 | 0.4182 | No |
| 142 | MRPL11 | MRPL11 Entrez, Source | mitochondrial ribosomal protein L11 | 4485 | 0.049 | 0.4174 | No |
| 143 | MRPL44 | MRPL44 Entrez, Source | mitochondrial ribosomal protein L44 | 4514 | 0.048 | 0.4165 | No |
| 144 | DAP3 | DAP3 Entrez, Source | death associated protein 3 | 4590 | 0.047 | 0.4119 | No |
| 145 | MPST | MPST Entrez, Source | mercaptopyruvate sulfurtransferase | 4713 | 0.044 | 0.4038 | No |
| 146 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 4772 | 0.042 | 0.4004 | No |
| 147 | NR3C1 | NR3C1 Entrez, Source | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 4791 | 0.042 | 0.4001 | No |
| 148 | ACP6 | ACP6 Entrez, Source | acid phosphatase 6, lysophosphatidic | 4817 | 0.042 | 0.3993 | No |
| 149 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 4861 | 0.041 | 0.3971 | No |
| 150 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 4966 | 0.039 | 0.3901 | No |
| 151 | ATP5D | ATP5D Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 5013 | 0.038 | 0.3876 | No |
| 152 | AK3 | AK3 Entrez, Source | adenylate kinase 3 | 5082 | 0.036 | 0.3834 | No |
| 153 | CYP27B1 | CYP27B1 Entrez, Source | cytochrome P450, family 27, subfamily B, polypeptide 1 | 5248 | 0.033 | 0.3716 | No |
| 154 | MRPL34 | MRPL34 Entrez, Source | mitochondrial ribosomal protein L34 | 5252 | 0.033 | 0.3722 | No |
| 155 | SUCLG1 | SUCLG1 Entrez, Source | succinate-CoA ligase, GDP-forming, alpha subunit | 5260 | 0.033 | 0.3725 | No |
| 156 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 5320 | 0.032 | 0.3689 | No |
| 157 | SLC1A3 | SLC1A3 Entrez, Source | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 5362 | 0.031 | 0.3665 | No |
| 158 | COX4I1 | COX4I1 Entrez, Source | cytochrome c oxidase subunit IV isoform 1 | 5380 | 0.031 | 0.3660 | No |
| 159 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5395 | 0.030 | 0.3657 | No |
| 160 | MRPL19 | MRPL19 Entrez, Source | mitochondrial ribosomal protein L19 | 5694 | 0.024 | 0.3436 | No |
| 161 | CRAT | CRAT Entrez, Source | carnitine acetyltransferase | 5716 | 0.023 | 0.3426 | No |
| 162 | MRPL17 | MRPL17 Entrez, Source | mitochondrial ribosomal protein L17 | 5720 | 0.023 | 0.3430 | No |
| 163 | ATP5B | ATP5B Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | 5777 | 0.022 | 0.3393 | No |
| 164 | COX5A | COX5A Entrez, Source | cytochrome c oxidase subunit Va | 5800 | 0.022 | 0.3382 | No |
| 165 | SUPV3L1 | SUPV3L1 Entrez, Source | suppressor of var1, 3-like 1 (S. cerevisiae) | 5802 | 0.022 | 0.3387 | No |
| 166 | PLA2G1B | PLA2G1B Entrez, Source | phospholipase A2, group IB (pancreas) | 6057 | 0.017 | 0.3197 | No |
| 167 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.3031 | No |
| 168 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 6292 | 0.012 | 0.3025 | No |
| 169 | MRPL9 | MRPL9 Entrez, Source | mitochondrial ribosomal protein L9 | 6314 | 0.012 | 0.3012 | No |
| 170 | CRY1 | CRY1 Entrez, Source | cryptochrome 1 (photolyase-like) | 6356 | 0.010 | 0.2984 | No |
| 171 | PMPCB | PMPCB Entrez, Source | peptidase (mitochondrial processing) beta | 6406 | 0.009 | 0.2949 | No |
| 172 | IDH3A | IDH3A Entrez, Source | isocitrate dehydrogenase 3 (NAD+) alpha | 6433 | 0.009 | 0.2931 | No |
| 173 | DIABLO | DIABLO Entrez, Source | diablo homolog (Drosophila) | 6449 | 0.009 | 0.2922 | No |
| 174 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 6477 | 0.008 | 0.2903 | No |
| 175 | NDUFA5 | NDUFA5 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa | 6553 | 0.007 | 0.2848 | No |
| 176 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 6598 | 0.006 | 0.2816 | No |
| 177 | CLN3 | CLN3 Entrez, Source | ceroid-lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 6686 | 0.004 | 0.2750 | No |
| 178 | NDUFB4 | NDUFB4 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa | 6690 | 0.004 | 0.2749 | No |
| 179 | C1QBP | C1QBP Entrez, Source | complement component 1, q subcomponent binding protein | 6697 | 0.004 | 0.2746 | No |
| 180 | MTIF2 | MTIF2 Entrez, Source | mitochondrial translational initiation factor 2 | 6712 | 0.004 | 0.2736 | No |
| 181 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 6721 | 0.004 | 0.2731 | No |
| 182 | HSPD1 | HSPD1 Entrez, Source | heat shock 60kDa protein 1 (chaperonin) | 6772 | 0.003 | 0.2693 | No |
| 183 | ABCB8 | ABCB8 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | 6773 | 0.003 | 0.2694 | No |
| 184 | MRPL13 | MRPL13 Entrez, Source | mitochondrial ribosomal protein L13 | 6822 | 0.002 | 0.2658 | No |
| 185 | COX6A1 | COX6A1 Entrez, Source | cytochrome c oxidase subunit VIa polypeptide 1 | 6867 | 0.001 | 0.2625 | No |
| 186 | IMMT | IMMT Entrez, Source | inner membrane protein, mitochondrial (mitofilin) | 6876 | 0.001 | 0.2619 | No |
| 187 | VDAC3 | VDAC3 Entrez, Source | voltage-dependent anion channel 3 | 7025 | -0.003 | 0.2506 | No |
| 188 | SSBP1 | SSBP1 Entrez, Source | single-stranded DNA binding protein 1 | 7057 | -0.003 | 0.2483 | No |
| 189 | TOMM20 | TOMM20 Entrez, Source | translocase of outer mitochondrial membrane 20 homolog (yeast) | 7072 | -0.004 | 0.2474 | No |
| 190 | MRPS18A | MRPS18A Entrez, Source | mitochondrial ribosomal protein S18A | 7193 | -0.006 | 0.2384 | No |
| 191 | MFN2 | MFN2 Entrez, Source | mitofusin 2 | 7196 | -0.006 | 0.2384 | No |
| 192 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 7234 | -0.007 | 0.2357 | No |
| 193 | NDUFA8 | NDUFA8 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa | 7295 | -0.008 | 0.2313 | No |
| 194 | MRPL2 | MRPL2 Entrez, Source | mitochondrial ribosomal protein L2 | 7336 | -0.009 | 0.2285 | No |
| 195 | DNM1L | DNM1L Entrez, Source | dynamin 1-like | 7354 | -0.009 | 0.2275 | No |
| 196 | MTERF | MTERF Entrez, Source | mitochondrial transcription termination factor | 7400 | -0.010 | 0.2243 | No |
| 197 | TIMM44 | TIMM44 Entrez, Source | translocase of inner mitochondrial membrane 44 homolog (yeast) | 7409 | -0.010 | 0.2239 | No |
| 198 | MRPL24 | MRPL24 Entrez, Source | mitochondrial ribosomal protein L24 | 7475 | -0.012 | 0.2193 | No |
| 199 | CS | CS Entrez, Source | citrate synthase | 7513 | -0.013 | 0.2168 | No |
| 200 | ATP5H | ATP5H Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | 7543 | -0.013 | 0.2149 | No |
| 201 | LYPLA2 | LYPLA2 Entrez, Source | lysophospholipase II | 7583 | -0.014 | 0.2123 | No |
| 202 | NME2 | NME2 Entrez, Source | non-metastatic cells 2, protein (NM23B) expressed in | 7595 | -0.014 | 0.2118 | No |
| 203 | CYP11B1 | CYP11B1 Entrez, Source | cytochrome P450, family 11, subfamily B, polypeptide 1 | 7700 | -0.017 | 0.2043 | No |
| 204 | TFAM | TFAM Entrez, Source | transcription factor A, mitochondrial | 7833 | -0.019 | 0.1947 | No |
| 205 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 7908 | -0.021 | 0.1896 | No |
| 206 | SLC1A1 | SLC1A1 Entrez, Source | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | 8024 | -0.024 | 0.1815 | No |
| 207 | TPO | TPO Entrez, Source | thyroid peroxidase | 8032 | -0.024 | 0.1816 | No |
| 208 | TIMM13 | TIMM13 Entrez, Source | translocase of inner mitochondrial membrane 13 homolog (yeast) | 8076 | -0.025 | 0.1789 | No |
| 209 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 8212 | -0.028 | 0.1693 | No |
| 210 | BCAT2 | BCAT2 Entrez, Source | branched chain aminotransferase 2, mitochondrial | 8232 | -0.028 | 0.1686 | No |
| 211 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 8404 | -0.032 | 0.1564 | No |
| 212 | CYBB | CYBB Entrez, Source | cytochrome b-245, beta polypeptide (chronic granulomatous disease) | 8513 | -0.035 | 0.1490 | No |
| 213 | BCL2L1 | BCL2L1 Entrez, Source | BCL2-like 1 | 8585 | -0.037 | 0.1446 | No |
| 214 | PRKCD | PRKCD Entrez, Source | protein kinase C, delta | 8690 | -0.039 | 0.1376 | No |
| 215 | CA5B | CA5B Entrez, Source | carbonic anhydrase VB, mitochondrial | 8760 | -0.040 | 0.1334 | No |
| 216 | SLC25A11 | SLC25A11 Entrez, Source | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | 8929 | -0.044 | 0.1217 | No |
| 217 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8995 | -0.046 | 0.1179 | No |
| 218 | TOMM22 | TOMM22 Entrez, Source | translocase of outer mitochondrial membrane 22 homolog (yeast) | 9085 | -0.048 | 0.1124 | No |
| 219 | UCP3 | UCP3 Entrez, Source | uncoupling protein 3 (mitochondrial, proton carrier) | 9230 | -0.052 | 0.1027 | No |
| 220 | TIMM22 | TIMM22 Entrez, Source | translocase of inner mitochondrial membrane 22 homolog (yeast) | 9516 | -0.060 | 0.0825 | No |
| 221 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 9534 | -0.061 | 0.0828 | No |
| 222 | ANXA7 | ANXA7 Entrez, Source | annexin A7 | 9552 | -0.061 | 0.0831 | No |
| 223 | YME1L1 | YME1L1 Entrez, Source | YME1-like 1 (S. cerevisiae) | 9701 | -0.065 | 0.0735 | No |
| 224 | CYP20A1 | CYP20A1 Entrez, Source | cytochrome P450, family 20, subfamily A, polypeptide 1 | 9929 | -0.073 | 0.0580 | No |
| 225 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 10075 | -0.077 | 0.0489 | No |
| 226 | BID | BID Entrez, Source | BH3 interacting domain death agonist | 10406 | -0.088 | 0.0260 | No |
| 227 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 10578 | -0.094 | 0.0154 | No |
| 228 | BAX | BAX Entrez, Source | BCL2-associated X protein | 10699 | -0.098 | 0.0088 | No |
| 229 | MRPL49 | MRPL49 Entrez, Source | mitochondrial ribosomal protein L49 | 10732 | -0.100 | 0.0089 | No |
| 230 | ATPIF1 | ATPIF1 Entrez, Source | ATPase inhibitory factor 1 | 10815 | -0.103 | 0.0054 | No |
| 231 | FDX1 | FDX1 Entrez, Source | ferredoxin 1 | 10969 | -0.110 | -0.0035 | No |
| 232 | ATP5S | ATP5S Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s (factor B) | 11182 | -0.120 | -0.0165 | No |
| 233 | CDC42BPB | CDC42BPB Entrez, Source | CDC42 binding protein kinase beta (DMPK-like) | 11232 | -0.123 | -0.0171 | No |
| 234 | BIK | BIK Entrez, Source | BCL2-interacting killer (apoptosis-inducing) | 11307 | -0.127 | -0.0194 | No |
| 235 | FIBP | FIBP Entrez, Source | fibroblast growth factor (acidic) intracellular binding protein | 11371 | -0.130 | -0.0209 | No |
| 236 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 11400 | -0.131 | -0.0196 | No |
| 237 | CASP8 | CASP8 Entrez, Source | caspase 8, apoptosis-related cysteine peptidase | 11401 | -0.131 | -0.0162 | No |
| 238 | APP | APP Entrez, Source | amyloid beta (A4) precursor protein (peptidase nexin-II, Alzheimer disease) | 11404 | -0.131 | -0.0129 | No |
| 239 | DNASE2 | DNASE2 Entrez, Source | deoxyribonuclease II, lysosomal | 11563 | -0.141 | -0.0214 | No |
| 240 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11690 | -0.149 | -0.0271 | No |
| 241 | CYP11A1 | CYP11A1 Entrez, Source | cytochrome P450, family 11, subfamily A, polypeptide 1 | 11705 | -0.150 | -0.0243 | No |
| 242 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 11723 | -0.150 | -0.0217 | No |
| 243 | MTCH1 | MTCH1 Entrez, Source | mitochondrial carrier homolog 1 (C. elegans) | 11859 | -0.161 | -0.0278 | No |
| 244 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 12237 | -0.197 | -0.0515 | No |
| 245 | BAK1 | BAK1 Entrez, Source | BCL2-antagonist/killer 1 | 12372 | -0.212 | -0.0562 | No |
| 246 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 12440 | -0.220 | -0.0556 | No |
| 247 | CROT | CROT Entrez, Source | carnitine O-octanoyltransferase | 12469 | -0.223 | -0.0519 | No |
| 248 | APAF1 | APAF1 Entrez, Source | apoptotic peptidase activating factor | 12510 | -0.230 | -0.0490 | No |
| 249 | POPDC2 | POPDC2 Entrez, Source | popeye domain containing 2 | 12600 | -0.245 | -0.0494 | No |
| 250 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12636 | -0.251 | -0.0456 | No |
| 251 | COX15 | COX15 Entrez, Source | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 12707 | -0.266 | -0.0440 | No |
| 252 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 12849 | -0.302 | -0.0469 | No |
| 253 | CYBA | CYBA Entrez, Source | cytochrome b-245, alpha polypeptide | 12989 | -0.359 | -0.0482 | No |
| 254 | MAP3K1 | MAP3K1 Entrez, Source | mitogen-activated protein kinase kinase kinase 1 | 13003 | -0.366 | -0.0397 | No |
| 255 | NGFRAP1 | NGFRAP1 Entrez, Source | nerve growth factor receptor (TNFRSF16) associated protein 1 | 13127 | -0.447 | -0.0375 | No |
| 256 | CASP2 | CASP2 Entrez, Source | caspase 2, apoptosis-related cysteine peptidase (neural precursor cell expressed, developmentally down-regulated 2) | 13202 | -0.565 | -0.0284 | No |
| 257 | LGALS3 | LGALS3 Entrez, Source | lectin, galactoside-binding, soluble, 3 (galectin 3) | 13253 | -0.689 | -0.0143 | No |
| 258 | UCP2 | UCP2 Entrez, Source | uncoupling protein 2 (mitochondrial, proton carrier) | 13284 | -0.808 | 0.0044 | No |