Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
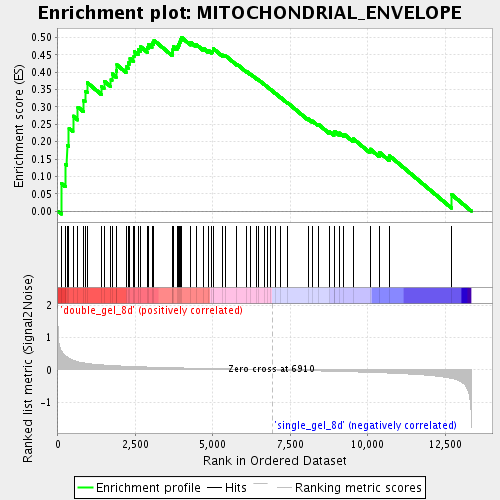
| GeneSet | MITOCHONDRIAL_ENVELOPE |
| Enrichment Score (ES) | 0.500882 |
| Normalized Enrichment Score (NES) | 1.7435395 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022130007 |
| FWER p-Value | 0.987 |

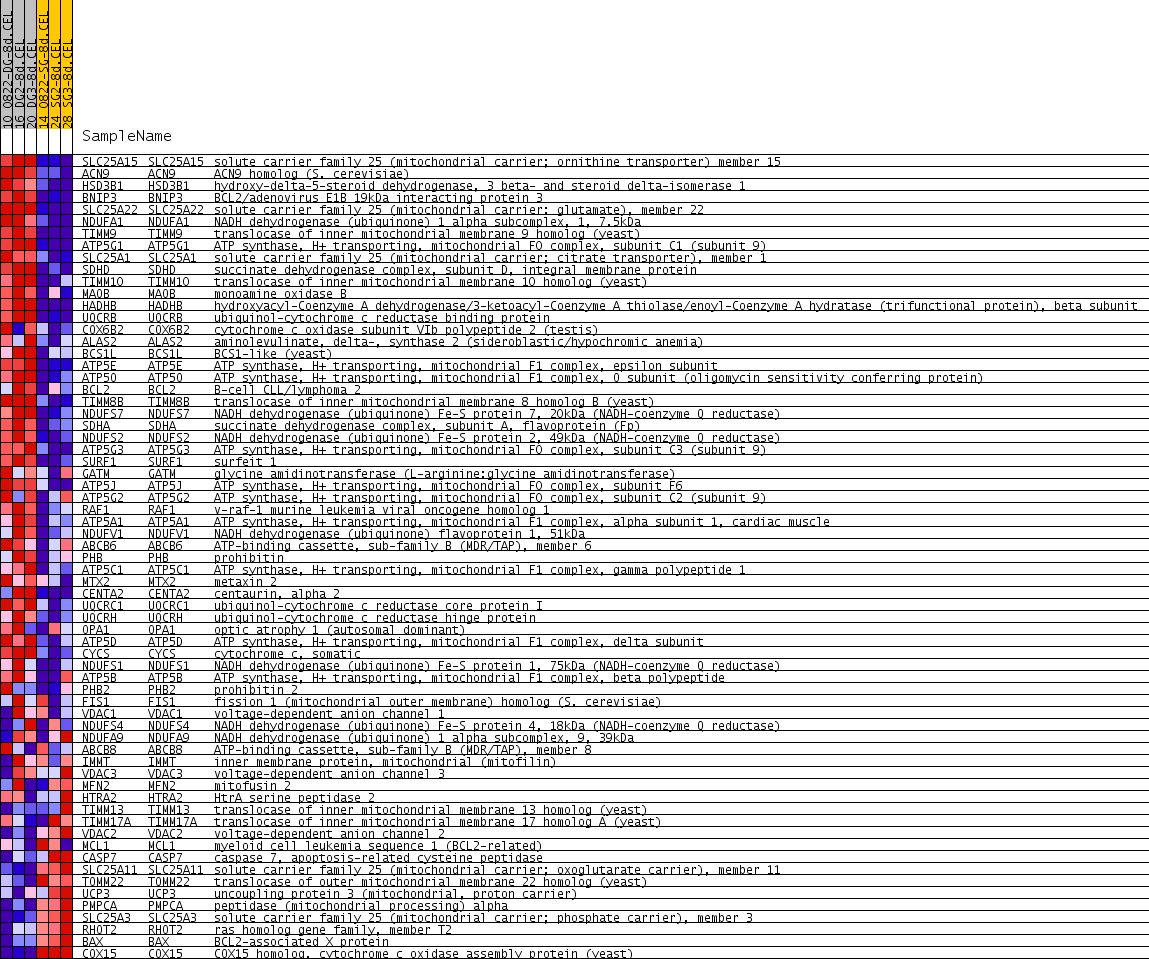
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 113 | 0.597 | 0.0797 | Yes |
| 2 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 247 | 0.438 | 0.1344 | Yes |
| 3 | HSD3B1 | HSD3B1 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | 293 | 0.402 | 0.1904 | Yes |
| 4 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 356 | 0.360 | 0.2389 | Yes |
| 5 | SLC25A22 | SLC25A22 Entrez, Source | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 488 | 0.305 | 0.2741 | Yes |
| 6 | NDUFA1 | NDUFA1 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa | 644 | 0.254 | 0.2999 | Yes |
| 7 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 828 | 0.216 | 0.3181 | Yes |
| 8 | ATP5G1 | ATP5G1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) | 878 | 0.210 | 0.3456 | Yes |
| 9 | SLC25A1 | SLC25A1 Entrez, Source | solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 | 946 | 0.200 | 0.3702 | Yes |
| 10 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 1404 | 0.157 | 0.3589 | Yes |
| 11 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 1491 | 0.150 | 0.3747 | Yes |
| 12 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1712 | 0.137 | 0.3783 | Yes |
| 13 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1747 | 0.134 | 0.3956 | Yes |
| 14 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 1880 | 0.127 | 0.4045 | Yes |
| 15 | COX6B2 | COX6B2 Entrez, Source | cytochrome c oxidase subunit VIb polypeptide 2 (testis) | 1903 | 0.126 | 0.4215 | Yes |
| 16 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 2203 | 0.114 | 0.4159 | Yes |
| 17 | BCS1L | BCS1L Entrez, Source | BCS1-like (yeast) | 2272 | 0.111 | 0.4272 | Yes |
| 18 | ATP5E | ATP5E Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | 2324 | 0.109 | 0.4395 | Yes |
| 19 | ATP5O | ATP5O Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) | 2450 | 0.105 | 0.4456 | Yes |
| 20 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 2467 | 0.104 | 0.4598 | Yes |
| 21 | TIMM8B | TIMM8B Entrez, Source | translocase of inner mitochondrial membrane 8 homolog B (yeast) | 2592 | 0.100 | 0.4652 | Yes |
| 22 | NDUFS7 | NDUFS7 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | 2674 | 0.096 | 0.4734 | Yes |
| 23 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2886 | 0.088 | 0.4705 | Yes |
| 24 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 2926 | 0.087 | 0.4805 | Yes |
| 25 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 3038 | 0.084 | 0.4846 | Yes |
| 26 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 3098 | 0.082 | 0.4922 | Yes |
| 27 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 3691 | 0.067 | 0.4574 | Yes |
| 28 | ATP5J | ATP5J Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 3709 | 0.066 | 0.4659 | Yes |
| 29 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 3731 | 0.066 | 0.4741 | Yes |
| 30 | RAF1 | RAF1 Entrez, Source | v-raf-1 murine leukemia viral oncogene homolog 1 | 3847 | 0.063 | 0.4747 | Yes |
| 31 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 3895 | 0.062 | 0.4804 | Yes |
| 32 | NDUFV1 | NDUFV1 Entrez, Source | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | 3926 | 0.061 | 0.4872 | Yes |
| 33 | ABCB6 | ABCB6 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 6 | 3958 | 0.061 | 0.4938 | Yes |
| 34 | PHB | PHB Entrez, Source | prohibitin | 3983 | 0.060 | 0.5009 | Yes |
| 35 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 4283 | 0.053 | 0.4862 | No |
| 36 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 4456 | 0.049 | 0.4806 | No |
| 37 | CENTA2 | CENTA2 Entrez, Source | centaurin, alpha 2 | 4710 | 0.044 | 0.4680 | No |
| 38 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 4861 | 0.041 | 0.4627 | No |
| 39 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 4966 | 0.039 | 0.4605 | No |
| 40 | OPA1 | OPA1 Entrez, Source | optic atrophy 1 (autosomal dominant) | 5005 | 0.038 | 0.4633 | No |
| 41 | ATP5D | ATP5D Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 5013 | 0.038 | 0.4683 | No |
| 42 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 5320 | 0.032 | 0.4499 | No |
| 43 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5395 | 0.030 | 0.4488 | No |
| 44 | ATP5B | ATP5B Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | 5777 | 0.022 | 0.4234 | No |
| 45 | PHB2 | PHB2 Entrez, Source | prohibitin 2 | 6088 | 0.016 | 0.4025 | No |
| 46 | FIS1 | FIS1 Entrez, Source | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | 6215 | 0.014 | 0.3951 | No |
| 47 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 6421 | 0.009 | 0.3810 | No |
| 48 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 6477 | 0.008 | 0.3780 | No |
| 49 | NDUFA9 | NDUFA9 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 6665 | 0.005 | 0.3647 | No |
| 50 | ABCB8 | ABCB8 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | 6773 | 0.003 | 0.3570 | No |
| 51 | IMMT | IMMT Entrez, Source | inner membrane protein, mitochondrial (mitofilin) | 6876 | 0.001 | 0.3495 | No |
| 52 | VDAC3 | VDAC3 Entrez, Source | voltage-dependent anion channel 3 | 7025 | -0.003 | 0.3387 | No |
| 53 | MFN2 | MFN2 Entrez, Source | mitofusin 2 | 7196 | -0.006 | 0.3268 | No |
| 54 | HTRA2 | HTRA2 Entrez, Source | HtrA serine peptidase 2 | 7406 | -0.010 | 0.3126 | No |
| 55 | TIMM13 | TIMM13 Entrez, Source | translocase of inner mitochondrial membrane 13 homolog (yeast) | 8076 | -0.025 | 0.2659 | No |
| 56 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 8212 | -0.028 | 0.2598 | No |
| 57 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 8404 | -0.032 | 0.2502 | No |
| 58 | MCL1 | MCL1 Entrez, Source | myeloid cell leukemia sequence 1 (BCL2-related) | 8765 | -0.040 | 0.2290 | No |
| 59 | CASP7 | CASP7 Entrez, Source | caspase 7, apoptosis-related cysteine peptidase | 8912 | -0.044 | 0.2245 | No |
| 60 | SLC25A11 | SLC25A11 Entrez, Source | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | 8929 | -0.044 | 0.2299 | No |
| 61 | TOMM22 | TOMM22 Entrez, Source | translocase of outer mitochondrial membrane 22 homolog (yeast) | 9085 | -0.048 | 0.2253 | No |
| 62 | UCP3 | UCP3 Entrez, Source | uncoupling protein 3 (mitochondrial, proton carrier) | 9230 | -0.052 | 0.2222 | No |
| 63 | PMPCA | PMPCA Entrez, Source | peptidase (mitochondrial processing) alpha | 9532 | -0.061 | 0.2085 | No |
| 64 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 10075 | -0.077 | 0.1790 | No |
| 65 | RHOT2 | RHOT2 Entrez, Source | ras homolog gene family, member T2 | 10381 | -0.088 | 0.1690 | No |
| 66 | BAX | BAX Entrez, Source | BCL2-associated X protein | 10699 | -0.098 | 0.1597 | No |
| 67 | COX15 | COX15 Entrez, Source | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 12707 | -0.266 | 0.0478 | No |