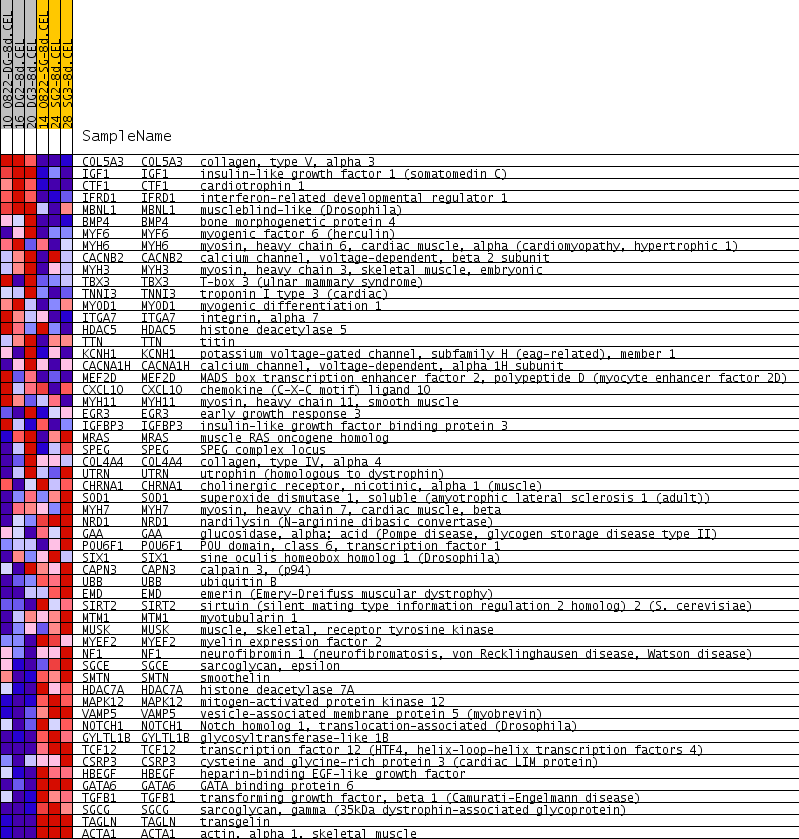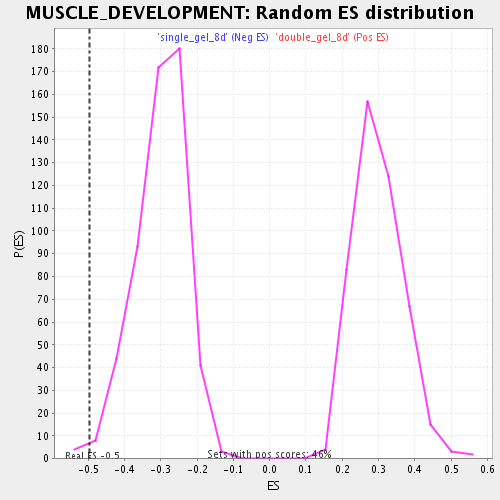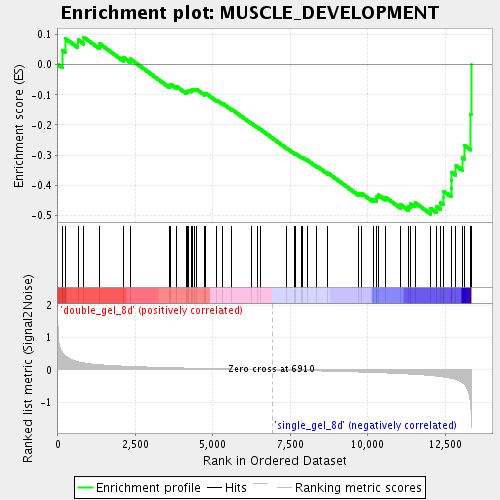
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | MUSCLE_DEVELOPMENT |
| Enrichment Score (ES) | -0.4956244 |
| Normalized Enrichment Score (NES) | -1.6432235 |
| Nominal p-value | 0.0091743115 |
| FDR q-value | 0.099971585 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | COL5A3 | COL5A3 Entrez, Source | collagen, type V, alpha 3 | 139 | 0.553 | 0.0475 | No |
| 2 | IGF1 | IGF1 Entrez, Source | insulin-like growth factor 1 (somatomedin C) | 236 | 0.445 | 0.0869 | No |
| 3 | CTF1 | CTF1 Entrez, Source | cardiotrophin 1 | 648 | 0.252 | 0.0824 | No |
| 4 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 837 | 0.215 | 0.0909 | No |
| 5 | MBNL1 | MBNL1 Entrez, Source | muscleblind-like (Drosophila) | 1343 | 0.161 | 0.0697 | No |
| 6 | BMP4 | BMP4 Entrez, Source | bone morphogenetic protein 4 | 2116 | 0.117 | 0.0239 | No |
| 7 | MYF6 | MYF6 Entrez, Source | myogenic factor 6 (herculin) | 2334 | 0.109 | 0.0190 | No |
| 8 | MYH6 | MYH6 Entrez, Source | myosin, heavy chain 6, cardiac muscle, alpha (cardiomyopathy, hypertrophic 1) | 3603 | 0.069 | -0.0693 | No |
| 9 | CACNB2 | CACNB2 Entrez, Source | calcium channel, voltage-dependent, beta 2 subunit | 3644 | 0.068 | -0.0652 | No |
| 10 | MYH3 | MYH3 Entrez, Source | myosin, heavy chain 3, skeletal muscle, embryonic | 3820 | 0.064 | -0.0717 | No |
| 11 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 4143 | 0.056 | -0.0900 | No |
| 12 | TNNI3 | TNNI3 Entrez, Source | troponin I type 3 (cardiac) | 4196 | 0.055 | -0.0882 | No |
| 13 | MYOD1 | MYOD1 Entrez, Source | myogenic differentiation 1 | 4224 | 0.054 | -0.0845 | No |
| 14 | ITGA7 | ITGA7 Entrez, Source | integrin, alpha 7 | 4309 | 0.053 | -0.0853 | No |
| 15 | HDAC5 | HDAC5 Entrez, Source | histone deacetylase 5 | 4333 | 0.052 | -0.0816 | No |
| 16 | TTN | TTN Entrez, Source | titin | 4401 | 0.050 | -0.0813 | No |
| 17 | KCNH1 | KCNH1 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 1 | 4463 | 0.049 | -0.0808 | No |
| 18 | CACNA1H | CACNA1H Entrez, Source | calcium channel, voltage-dependent, alpha 1H subunit | 4735 | 0.043 | -0.0967 | No |
| 19 | MEF2D | MEF2D Entrez, Source | MADS box transcription enhancer factor 2, polypeptide D (myocyte enhancer factor 2D) | 4769 | 0.042 | -0.0947 | No |
| 20 | CXCL10 | CXCL10 Entrez, Source | chemokine (C-X-C motif) ligand 10 | 5128 | 0.035 | -0.1180 | No |
| 21 | MYH11 | MYH11 Entrez, Source | myosin, heavy chain 11, smooth muscle | 5315 | 0.032 | -0.1286 | No |
| 22 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 5611 | 0.025 | -0.1482 | No |
| 23 | IGFBP3 | IGFBP3 Entrez, Source | insulin-like growth factor binding protein 3 | 6247 | 0.013 | -0.1946 | No |
| 24 | MRAS | MRAS Entrez, Source | muscle RAS oncogene homolog | 6436 | 0.009 | -0.2078 | No |
| 25 | SPEG | SPEG Entrez, Source | SPEG complex locus | 6531 | 0.007 | -0.2141 | No |
| 26 | COL4A4 | COL4A4 Entrez, Source | collagen, type IV, alpha 4 | 7382 | -0.010 | -0.2771 | No |
| 27 | UTRN | UTRN Entrez, Source | utrophin (homologous to dystrophin) | 7645 | -0.015 | -0.2952 | No |
| 28 | CHRNA1 | CHRNA1 Entrez, Source | cholinergic receptor, nicotinic, alpha 1 (muscle) | 7663 | -0.016 | -0.2948 | No |
| 29 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7856 | -0.020 | -0.3072 | No |
| 30 | MYH7 | MYH7 Entrez, Source | myosin, heavy chain 7, cardiac muscle, beta | 7898 | -0.021 | -0.3080 | No |
| 31 | NRD1 | NRD1 Entrez, Source | nardilysin (N-arginine dibasic convertase) | 8039 | -0.024 | -0.3160 | No |
| 32 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 8354 | -0.031 | -0.3364 | No |
| 33 | POU6F1 | POU6F1 Entrez, Source | POU domain, class 6, transcription factor 1 | 8710 | -0.039 | -0.3590 | No |
| 34 | SIX1 | SIX1 Entrez, Source | sine oculis homeobox homolog 1 (Drosophila) | 9691 | -0.065 | -0.4260 | No |
| 35 | CAPN3 | CAPN3 Entrez, Source | calpain 3, (p94) | 9789 | -0.068 | -0.4262 | No |
| 36 | UBB | UBB Entrez, Source | ubiquitin B | 10179 | -0.081 | -0.4470 | No |
| 37 | EMD | EMD Entrez, Source | emerin (Emery-Dreifuss muscular dystrophy) | 10290 | -0.084 | -0.4464 | No |
| 38 | SIRT2 | SIRT2 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | 10295 | -0.085 | -0.4379 | No |
| 39 | MTM1 | MTM1 Entrez, Source | myotubularin 1 | 10339 | -0.086 | -0.4321 | No |
| 40 | MUSK | MUSK Entrez, Source | muscle, skeletal, receptor tyrosine kinase | 10568 | -0.094 | -0.4394 | No |
| 41 | MYEF2 | MYEF2 Entrez, Source | myelin expression factor 2 | 11057 | -0.115 | -0.4641 | No |
| 42 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 11321 | -0.127 | -0.4706 | No |
| 43 | SGCE | SGCE Entrez, Source | sarcoglycan, epsilon | 11390 | -0.131 | -0.4620 | No |
| 44 | SMTN | SMTN Entrez, Source | smoothelin | 11525 | -0.139 | -0.4575 | No |
| 45 | HDAC7A | HDAC7A Entrez, Source | histone deacetylase 7A | 12032 | -0.176 | -0.4772 | Yes |
| 46 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 12219 | -0.194 | -0.4709 | Yes |
| 47 | VAMP5 | VAMP5 Entrez, Source | vesicle-associated membrane protein 5 (myobrevin) | 12345 | -0.209 | -0.4584 | Yes |
| 48 | NOTCH1 | NOTCH1 Entrez, Source | Notch homolog 1, translocation-associated (Drosophila) | 12433 | -0.219 | -0.4420 | Yes |
| 49 | GYLTL1B | GYLTL1B Entrez, Source | glycosyltransferase-like 1B | 12436 | -0.220 | -0.4191 | Yes |
| 50 | TCF12 | TCF12 Entrez, Source | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | 12690 | -0.262 | -0.4107 | Yes |
| 51 | CSRP3 | CSRP3 Entrez, Source | cysteine and glycine-rich protein 3 (cardiac LIM protein) | 12703 | -0.265 | -0.3838 | Yes |
| 52 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 12714 | -0.267 | -0.3566 | Yes |
| 53 | GATA6 | GATA6 Entrez, Source | GATA binding protein 6 | 12847 | -0.302 | -0.3348 | Yes |
| 54 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 13047 | -0.395 | -0.3084 | Yes |
| 55 | SGCG | SGCG Entrez, Source | sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) | 13113 | -0.440 | -0.2672 | Yes |
| 56 | TAGLN | TAGLN Entrez, Source | transgelin | 13326 | -1.131 | -0.1646 | Yes |
| 57 | ACTA1 | ACTA1 Entrez, Source | actin, alpha 1, skeletal muscle | 13340 | -1.581 | 0.0002 | Yes |