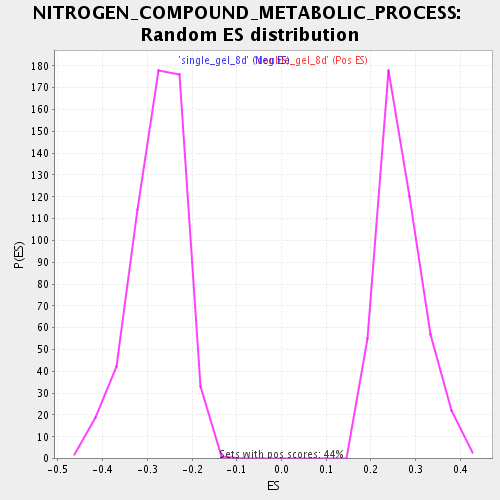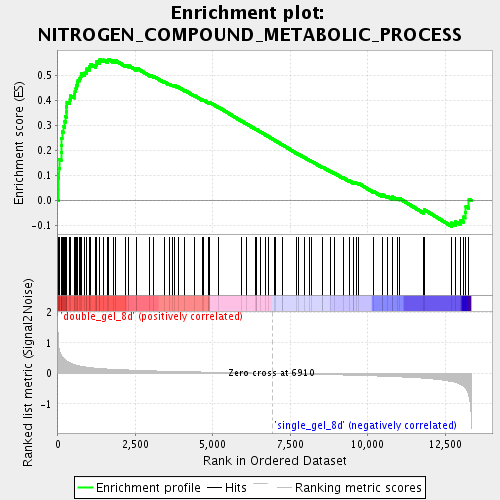
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | NITROGEN_COMPOUND_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.56428427 |
| Normalized Enrichment Score (NES) | 2.1104662 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.6288786E-4 |
| FWER p-Value | 0.023 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 16 | 0.933 | 0.0449 | Yes |
| 2 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 17 | 0.930 | 0.0909 | Yes |
| 3 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 35 | 0.815 | 0.1299 | Yes |
| 4 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 56 | 0.731 | 0.1645 | Yes |
| 5 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 104 | 0.612 | 0.1912 | Yes |
| 6 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 113 | 0.597 | 0.2201 | Yes |
| 7 | ASL | ASL Entrez, Source | argininosuccinate lyase | 122 | 0.586 | 0.2485 | Yes |
| 8 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 135 | 0.556 | 0.2751 | Yes |
| 9 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 192 | 0.482 | 0.2947 | Yes |
| 10 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 223 | 0.456 | 0.3150 | Yes |
| 11 | ARG1 | ARG1 Entrez, Source | arginase, liver | 246 | 0.438 | 0.3350 | Yes |
| 12 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 271 | 0.419 | 0.3539 | Yes |
| 13 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 277 | 0.416 | 0.3741 | Yes |
| 14 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 291 | 0.403 | 0.3931 | Yes |
| 15 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 389 | 0.344 | 0.4027 | Yes |
| 16 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 411 | 0.336 | 0.4178 | Yes |
| 17 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 524 | 0.292 | 0.4238 | Yes |
| 18 | SULT1C2 | SULT1C2 Entrez, Source | sulfotransferase family, cytosolic, 1C, member 2 | 550 | 0.282 | 0.4358 | Yes |
| 19 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 580 | 0.273 | 0.4471 | Yes |
| 20 | ASS1 | ASS1 Entrez, Source | argininosuccinate synthetase 1 | 598 | 0.267 | 0.4591 | Yes |
| 21 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 623 | 0.260 | 0.4701 | Yes |
| 22 | COLQ | COLQ Entrez, Source | collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | 646 | 0.252 | 0.4809 | Yes |
| 23 | SLC35A3 | SLC35A3 Entrez, Source | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 | 697 | 0.239 | 0.4890 | Yes |
| 24 | SULT1B1 | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 743 | 0.229 | 0.4969 | Yes |
| 25 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 753 | 0.227 | 0.5075 | Yes |
| 26 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 870 | 0.211 | 0.5091 | Yes |
| 27 | GCHFR | GCHFR Entrez, Source | GTP cyclohydrolase I feedback regulator | 921 | 0.204 | 0.5154 | Yes |
| 28 | TST | TST Entrez, Source | thiosulfate sulfurtransferase (rhodanese) | 926 | 0.203 | 0.5252 | Yes |
| 29 | SULT1A1 | SULT1A1 Entrez, Source | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 | 1008 | 0.192 | 0.5285 | Yes |
| 30 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 1027 | 0.189 | 0.5366 | Yes |
| 31 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 1046 | 0.187 | 0.5445 | Yes |
| 32 | SDS | SDS Entrez, Source | serine dehydratase | 1222 | 0.169 | 0.5396 | Yes |
| 33 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 1252 | 0.167 | 0.5457 | Yes |
| 34 | HDC | HDC Entrez, Source | histidine decarboxylase | 1256 | 0.167 | 0.5537 | Yes |
| 35 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 1345 | 0.161 | 0.5550 | Yes |
| 36 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 1355 | 0.160 | 0.5623 | Yes |
| 37 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 1469 | 0.151 | 0.5612 | Yes |
| 38 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 1598 | 0.143 | 0.5586 | Yes |
| 39 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 1617 | 0.142 | 0.5643 | Yes |
| 40 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 1785 | 0.132 | 0.5582 | No |
| 41 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 1864 | 0.128 | 0.5586 | No |
| 42 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2183 | 0.115 | 0.5403 | No |
| 43 | GALNT5 | GALNT5 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | 2282 | 0.111 | 0.5384 | No |
| 44 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 2527 | 0.102 | 0.5250 | No |
| 45 | SPR | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 2551 | 0.101 | 0.5283 | No |
| 46 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 2968 | 0.086 | 0.5011 | No |
| 47 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 3082 | 0.082 | 0.4966 | No |
| 48 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 3436 | 0.073 | 0.4735 | No |
| 49 | GSS | GSS Entrez, Source | glutathione synthetase | 3596 | 0.069 | 0.4649 | No |
| 50 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 3691 | 0.067 | 0.4611 | No |
| 51 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 3774 | 0.065 | 0.4581 | No |
| 52 | AMT | AMT Entrez, Source | aminomethyltransferase | 3875 | 0.063 | 0.4537 | No |
| 53 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 4100 | 0.057 | 0.4396 | No |
| 54 | XYLT1 | XYLT1 Entrez, Source | xylosyltransferase I | 4421 | 0.050 | 0.4179 | No |
| 55 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 4665 | 0.045 | 0.4017 | No |
| 56 | MPST | MPST Entrez, Source | mercaptopyruvate sulfurtransferase | 4713 | 0.044 | 0.4004 | No |
| 57 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 4870 | 0.040 | 0.3906 | No |
| 58 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 4874 | 0.040 | 0.3923 | No |
| 59 | B4GALT7 | B4GALT7 Entrez, Source | xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) | 4905 | 0.040 | 0.3920 | No |
| 60 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 5183 | 0.034 | 0.3728 | No |
| 61 | SLC5A7 | SLC5A7 Entrez, Source | solute carrier family 5 (choline transporter), member 7 | 5926 | 0.020 | 0.3177 | No |
| 62 | CHST12 | CHST12 Entrez, Source | carbohydrate (chondroitin 4) sulfotransferase 12 | 6090 | 0.016 | 0.3062 | No |
| 63 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 6381 | 0.010 | 0.2848 | No |
| 64 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 6397 | 0.010 | 0.2842 | No |
| 65 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 6420 | 0.009 | 0.2830 | No |
| 66 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 6525 | 0.007 | 0.2755 | No |
| 67 | NQO1 | NQO1 Entrez, Source | NAD(P)H dehydrogenase, quinone 1 | 6707 | 0.004 | 0.2620 | No |
| 68 | CHIA | CHIA Entrez, Source | chitinase, acidic | 6796 | 0.002 | 0.2555 | No |
| 69 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 6980 | -0.002 | 0.2417 | No |
| 70 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 7015 | -0.002 | 0.2393 | No |
| 71 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 7234 | -0.007 | 0.2232 | No |
| 72 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 7238 | -0.007 | 0.2233 | No |
| 73 | DHPS | DHPS Entrez, Source | deoxyhypusine synthase | 7692 | -0.016 | 0.1899 | No |
| 74 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 7757 | -0.018 | 0.1859 | No |
| 75 | NAGK | NAGK Entrez, Source | N-acetylglucosamine kinase | 7948 | -0.022 | 0.1727 | No |
| 76 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 8124 | -0.026 | 0.1608 | No |
| 77 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 8177 | -0.027 | 0.1582 | No |
| 78 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 8542 | -0.036 | 0.1324 | No |
| 79 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 8548 | -0.036 | 0.1338 | No |
| 80 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 8796 | -0.041 | 0.1172 | No |
| 81 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 8939 | -0.045 | 0.1087 | No |
| 82 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 9206 | -0.051 | 0.0911 | No |
| 83 | OAZ1 | OAZ1 Entrez, Source | ornithine decarboxylase antizyme 1 | 9397 | -0.057 | 0.0796 | No |
| 84 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 9551 | -0.061 | 0.0710 | No |
| 85 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 9553 | -0.061 | 0.0740 | No |
| 86 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 9642 | -0.064 | 0.0705 | No |
| 87 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 9713 | -0.066 | 0.0684 | No |
| 88 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 10171 | -0.080 | 0.0379 | No |
| 89 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 10462 | -0.090 | 0.0205 | No |
| 90 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10469 | -0.091 | 0.0245 | No |
| 91 | HSP90AA1 | HSP90AA1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class A member 1 | 10648 | -0.097 | 0.0158 | No |
| 92 | HSP90AB1 | HSP90AB1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class B member 1 | 10785 | -0.102 | 0.0106 | No |
| 93 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 10791 | -0.102 | 0.0153 | No |
| 94 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 10959 | -0.110 | 0.0081 | No |
| 95 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 11033 | -0.113 | 0.0082 | No |
| 96 | AKT1 | AKT1 Entrez, Source | v-akt murine thymoma viral oncogene homolog 1 | 11801 | -0.156 | -0.0420 | No |
| 97 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 11815 | -0.158 | -0.0352 | No |
| 98 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12700 | -0.264 | -0.0889 | No |
| 99 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12832 | -0.296 | -0.0842 | No |
| 100 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 12983 | -0.354 | -0.0780 | No |
| 101 | SMS | SMS Entrez, Source | spermine synthase | 13083 | -0.414 | -0.0650 | No |
| 102 | ST3GAL2 | ST3GAL2 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 13139 | -0.459 | -0.0464 | No |
| 103 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13167 | -0.505 | -0.0235 | No |
| 104 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 13266 | -0.741 | 0.0057 | No |