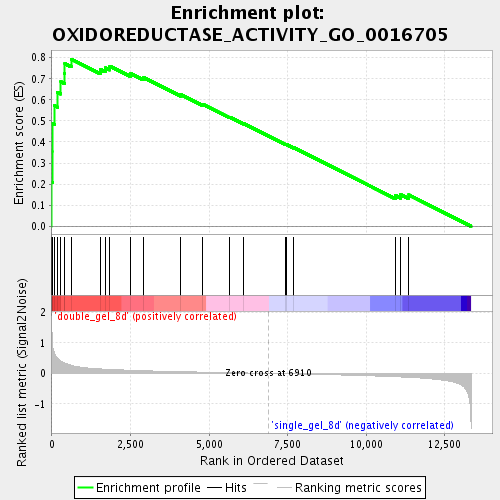
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | OXIDOREDUCTASE_ACTIVITY_GO_0016705 |
| Enrichment Score (ES) | 0.7933474 |
| Normalized Enrichment Score (NES) | 2.2325566 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.0418995E-5 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CYP17A1 | CYP17A1 Entrez, Source | cytochrome P450, family 17, subfamily A, polypeptide 1 | 1 | 1.475 | 0.2090 | Yes |
| 2 | CYP8B1 | CYP8B1 Entrez, Source | cytochrome P450, family 8, subfamily B, polypeptide 1 | 14 | 1.030 | 0.3541 | Yes |
| 3 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 16 | 0.933 | 0.4862 | Yes |
| 4 | SCD | SCD Entrez, Source | stearoyl-CoA desaturase (delta-9-desaturase) | 84 | 0.641 | 0.5721 | Yes |
| 5 | KMO | KMO Entrez, Source | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 174 | 0.497 | 0.6359 | Yes |
| 6 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 284 | 0.409 | 0.6857 | Yes |
| 7 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 401 | 0.338 | 0.7249 | Yes |
| 8 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 411 | 0.336 | 0.7718 | Yes |
| 9 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 617 | 0.261 | 0.7933 | Yes |
| 10 | HMOX2 | HMOX2 Entrez, Source | heme oxygenase (decycling) 2 | 1546 | 0.146 | 0.7444 | No |
| 11 | EGLN1 | EGLN1 Entrez, Source | egl nine homolog 1 (C. elegans) | 1704 | 0.137 | 0.7520 | No |
| 12 | NOS3 | NOS3 Entrez, Source | nitric oxide synthase 3 (endothelial cell) | 1840 | 0.129 | 0.7601 | No |
| 13 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 2499 | 0.103 | 0.7253 | No |
| 14 | CYP19A1 | CYP19A1 Entrez, Source | cytochrome P450, family 19, subfamily A, polypeptide 1 | 2909 | 0.088 | 0.7070 | No |
| 15 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 4100 | 0.057 | 0.6258 | No |
| 16 | NOS1 | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 4800 | 0.042 | 0.5792 | No |
| 17 | CYP26A1 | CYP26A1 Entrez, Source | cytochrome P450, family 26, subfamily A, polypeptide 1 | 5664 | 0.024 | 0.5179 | No |
| 18 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 6103 | 0.016 | 0.4873 | No |
| 19 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 7432 | -0.011 | 0.3891 | No |
| 20 | TH | TH Entrez, Source | tyrosine hydroxylase | 7455 | -0.011 | 0.3891 | No |
| 21 | CYP11B1 | CYP11B1 Entrez, Source | cytochrome P450, family 11, subfamily B, polypeptide 1 | 7700 | -0.017 | 0.3731 | No |
| 22 | PLOD3 | PLOD3 Entrez, Source | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | 10940 | -0.109 | 0.1453 | No |
| 23 | P4HA1 | P4HA1 Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I | 11102 | -0.117 | 0.1498 | No |
| 24 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 11363 | -0.129 | 0.1486 | No |