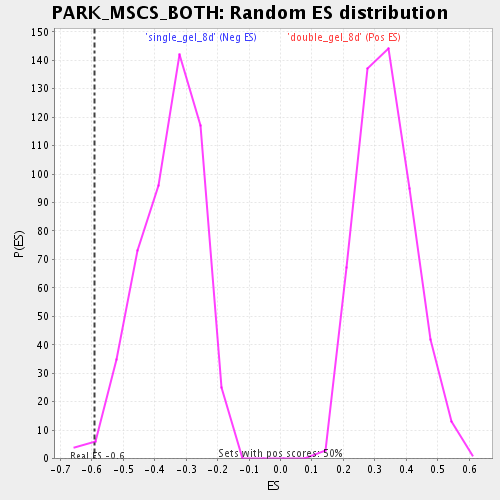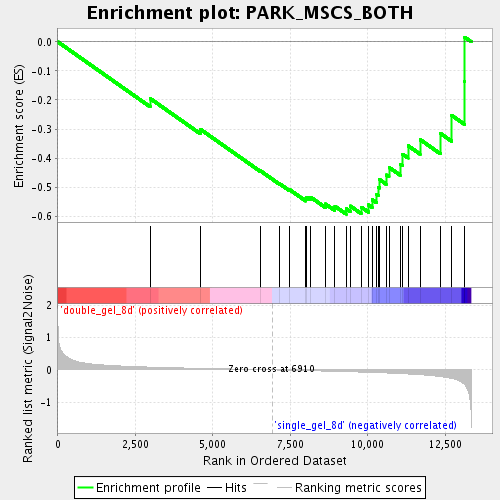
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | PARK_MSCS_BOTH |
| Enrichment Score (ES) | -0.5928782 |
| Normalized Enrichment Score (NES) | -1.6919833 |
| Nominal p-value | 0.012048192 |
| FDR q-value | 0.07786243 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 2979 | 0.086 | -0.1950 | No |
| 2 | RAMP1 | RAMP1 Entrez, Source | receptor (calcitonin) activity modifying protein 1 | 4600 | 0.046 | -0.3011 | No |
| 3 | PSMB2 | PSMB2 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 2 | 6524 | 0.007 | -0.4430 | No |
| 4 | ICAM1 | ICAM1 Entrez, Source | intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 7160 | -0.005 | -0.4889 | No |
| 5 | P2RY4 | P2RY4 Entrez, Source | pyrimidinergic receptor P2Y, G-protein coupled, 4 | 7469 | -0.012 | -0.5081 | No |
| 6 | PPIA | PPIA Entrez, Source | peptidylprolyl isomerase A (cyclophilin A) | 8000 | -0.023 | -0.5400 | No |
| 7 | TSN | TSN Entrez, Source | translin | 8029 | -0.024 | -0.5341 | No |
| 8 | SKP1A | SKP1A Entrez, Source | S-phase kinase-associated protein 1A (p19A) | 8149 | -0.026 | -0.5341 | No |
| 9 | SORBS3 | SORBS3 Entrez, Source | sorbin and SH3 domain containing 3 | 8634 | -0.037 | -0.5578 | No |
| 10 | RPSA | RPSA Entrez, Source | ribosomal protein SA | 8938 | -0.045 | -0.5656 | No |
| 11 | CNBP | CNBP Entrez, Source | CCHC-type zinc finger, nucleic acid binding protein | 9302 | -0.054 | -0.5746 | Yes |
| 12 | KPNB1 | KPNB1 Entrez, Source | karyopherin (importin) beta 1 | 9435 | -0.058 | -0.5649 | Yes |
| 13 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9788 | -0.068 | -0.5686 | Yes |
| 14 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 10026 | -0.075 | -0.5611 | Yes |
| 15 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 10143 | -0.079 | -0.5431 | Yes |
| 16 | H3F3B | H3F3B Entrez, Source | H3 histone, family 3B (H3.3B) | 10288 | -0.084 | -0.5256 | Yes |
| 17 | LMO2 | LMO2 Entrez, Source | LIM domain only 2 (rhombotin-like 1) | 10340 | -0.086 | -0.5004 | Yes |
| 18 | GDI2 | GDI2 Entrez, Source | GDP dissociation inhibitor 2 | 10378 | -0.087 | -0.4738 | Yes |
| 19 | RNF4 | RNF4 Entrez, Source | ring finger protein 4 | 10595 | -0.095 | -0.4581 | Yes |
| 20 | NAP1L4 | NAP1L4 Entrez, Source | nucleosome assembly protein 1-like 4 | 10686 | -0.098 | -0.4319 | Yes |
| 21 | ITM2B | ITM2B Entrez, Source | integral membrane protein 2B | 11047 | -0.114 | -0.4205 | Yes |
| 22 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 11117 | -0.118 | -0.3861 | Yes |
| 23 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 11301 | -0.126 | -0.3573 | Yes |
| 24 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11690 | -0.149 | -0.3363 | Yes |
| 25 | VAMP5 | VAMP5 Entrez, Source | vesicle-associated membrane protein 5 (myobrevin) | 12345 | -0.209 | -0.3151 | Yes |
| 26 | TAGLN2 | TAGLN2 Entrez, Source | transgelin 2 | 12708 | -0.266 | -0.2529 | Yes |
| 27 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 13120 | -0.443 | -0.1348 | Yes |
| 28 | RAMP2 | RAMP2 Entrez, Source | receptor (calcitonin) activity modifying protein 2 | 13129 | -0.450 | 0.0160 | Yes |