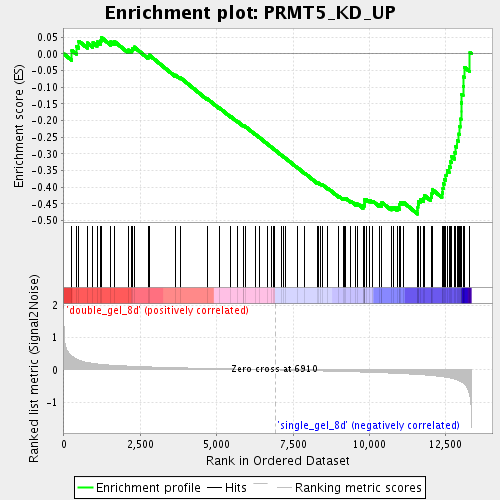
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | PRMT5_KD_UP |
| Enrichment Score (ES) | -0.48029447 |
| Normalized Enrichment Score (NES) | -1.7382729 |
| Nominal p-value | 0.0017793594 |
| FDR q-value | 0.06549089 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
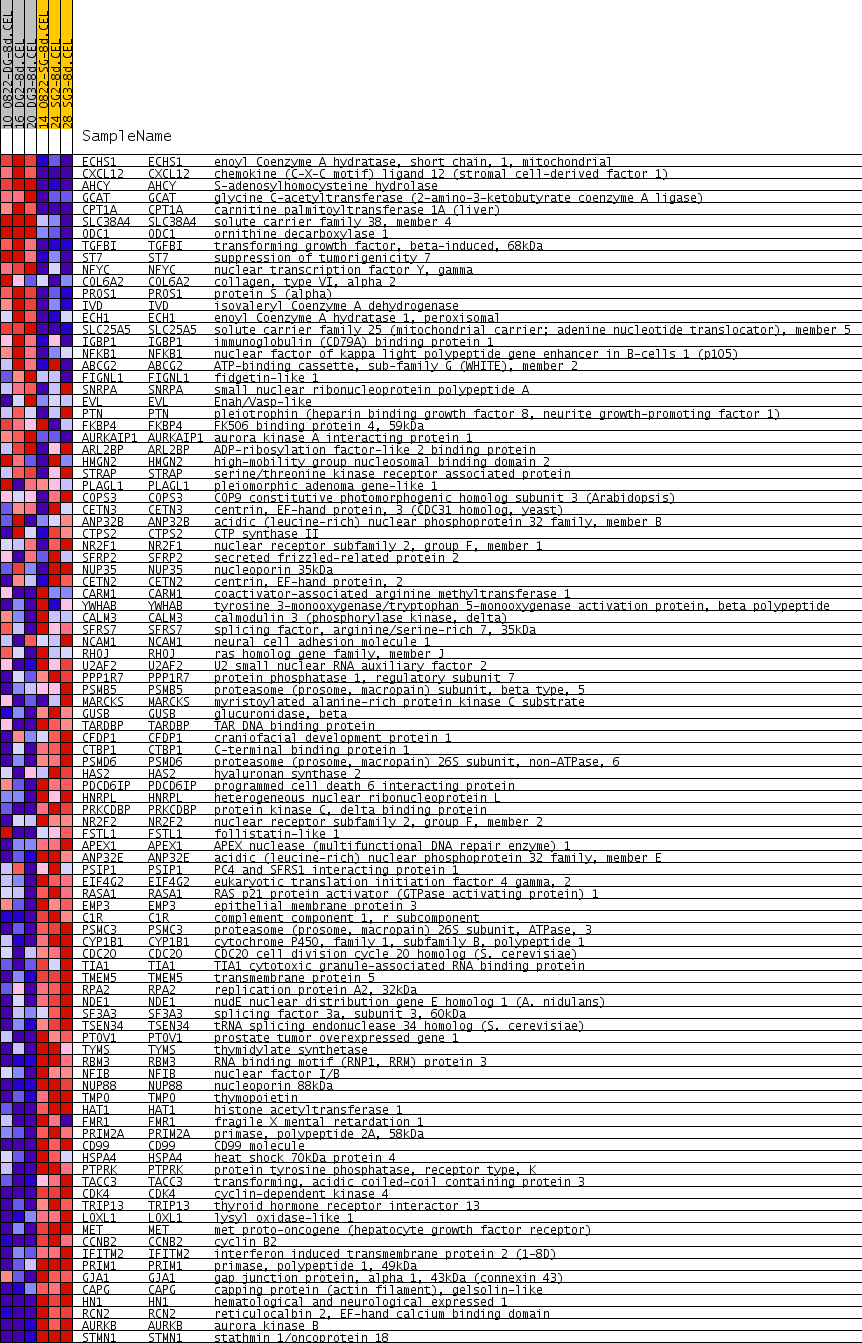
| 1 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.0102 | No |
| 2 | CXCL12 | CXCL12 Entrez, Source | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 407 | 0.337 | 0.0228 | No |
| 3 | AHCY | AHCY Entrez, Source | S-adenosylhomocysteine hydrolase | 485 | 0.309 | 0.0386 | No |
| 4 | GCAT | GCAT Entrez, Source | glycine C-acetyltransferase (2-amino-3-ketobutyrate coenzyme A ligase) | 767 | 0.225 | 0.0331 | No |
| 5 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 950 | 0.200 | 0.0333 | No |
| 6 | SLC38A4 | SLC38A4 Entrez, Source | solute carrier family 38, member 4 | 1084 | 0.184 | 0.0361 | No |
| 7 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 1205 | 0.170 | 0.0390 | No |
| 8 | TGFBI | TGFBI Entrez, Source | transforming growth factor, beta-induced, 68kDa | 1217 | 0.169 | 0.0500 | No |
| 9 | ST7 | ST7 Entrez, Source | suppression of tumorigenicity 7 | 1537 | 0.147 | 0.0361 | No |
| 10 | NFYC | NFYC Entrez, Source | nuclear transcription factor Y, gamma | 1659 | 0.139 | 0.0367 | No |
| 11 | COL6A2 | COL6A2 Entrez, Source | collagen, type VI, alpha 2 | 2097 | 0.118 | 0.0120 | No |
| 12 | PROS1 | PROS1 Entrez, Source | protein S (alpha) | 2217 | 0.114 | 0.0110 | No |
| 13 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 2248 | 0.113 | 0.0166 | No |
| 14 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 2294 | 0.111 | 0.0209 | No |
| 15 | SLC25A5 | SLC25A5 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 2756 | 0.094 | -0.0074 | No |
| 16 | IGBP1 | IGBP1 Entrez, Source | immunoglobulin (CD79A) binding protein 1 | 2784 | 0.092 | -0.0029 | No |
| 17 | NFKB1 | NFKB1 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 3646 | 0.068 | -0.0632 | No |
| 18 | ABCG2 | ABCG2 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 2 | 3807 | 0.064 | -0.0708 | No |
| 19 | FIGNL1 | FIGNL1 Entrez, Source | fidgetin-like 1 | 4683 | 0.044 | -0.1338 | No |
| 20 | SNRPA | SNRPA Entrez, Source | small nuclear ribonucleoprotein polypeptide A | 5088 | 0.036 | -0.1618 | No |
| 21 | EVL | EVL Entrez, Source | Enah/Vasp-like | 5460 | 0.029 | -0.1878 | No |
| 22 | PTN | PTN Entrez, Source | pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) | 5682 | 0.024 | -0.2028 | No |
| 23 | FKBP4 | FKBP4 Entrez, Source | FK506 binding protein 4, 59kDa | 5864 | 0.021 | -0.2150 | No |
| 24 | AURKAIP1 | AURKAIP1 Entrez, Source | aurora kinase A interacting protein 1 | 5880 | 0.021 | -0.2147 | No |
| 25 | ARL2BP | ARL2BP Entrez, Source | ADP-ribosylation factor-like 2 binding protein | 5950 | 0.019 | -0.2185 | No |
| 26 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 6264 | 0.013 | -0.2413 | No |
| 27 | STRAP | STRAP Entrez, Source | serine/threonine kinase receptor associated protein | 6387 | 0.010 | -0.2498 | No |
| 28 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 6668 | 0.005 | -0.2706 | No |
| 29 | COPS3 | COPS3 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 3 (Arabidopsis) | 6803 | 0.002 | -0.2806 | No |
| 30 | CETN3 | CETN3 Entrez, Source | centrin, EF-hand protein, 3 (CDC31 homolog, yeast) | 6856 | 0.001 | -0.2844 | No |
| 31 | ANP32B | ANP32B Entrez, Source | acidic (leucine-rich) nuclear phosphoprotein 32 family, member B | 6879 | 0.001 | -0.2860 | No |
| 32 | CTPS2 | CTPS2 Entrez, Source | CTP synthase II | 7130 | -0.005 | -0.3046 | No |
| 33 | NR2F1 | NR2F1 Entrez, Source | nuclear receptor subfamily 2, group F, member 1 | 7183 | -0.006 | -0.3081 | No |
| 34 | SFRP2 | SFRP2 Entrez, Source | secreted frizzled-related protein 2 | 7256 | -0.008 | -0.3130 | No |
| 35 | NUP35 | NUP35 Entrez, Source | nucleoporin 35kDa | 7632 | -0.015 | -0.3402 | No |
| 36 | CETN2 | CETN2 Entrez, Source | centrin, EF-hand protein, 2 | 7883 | -0.021 | -0.3577 | No |
| 37 | CARM1 | CARM1 Entrez, Source | coactivator-associated arginine methyltransferase 1 | 8288 | -0.030 | -0.3861 | No |
| 38 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 8325 | -0.031 | -0.3867 | No |
| 39 | CALM3 | CALM3 Entrez, Source | calmodulin 3 (phosphorylase kinase, delta) | 8407 | -0.032 | -0.3905 | No |
| 40 | SFRS7 | SFRS7 Entrez, Source | splicing factor, arginine/serine-rich 7, 35kDa | 8464 | -0.033 | -0.3924 | No |
| 41 | NCAM1 | NCAM1 Entrez, Source | neural cell adhesion molecule 1 | 8637 | -0.038 | -0.4028 | No |
| 42 | RHOJ | RHOJ Entrez, Source | ras homolog gene family, member J | 8997 | -0.046 | -0.4267 | No |
| 43 | U2AF2 | U2AF2 Entrez, Source | U2 small nuclear RNA auxiliary factor 2 | 9141 | -0.050 | -0.4340 | No |
| 44 | PPP1R7 | PPP1R7 Entrez, Source | protein phosphatase 1, regulatory subunit 7 | 9184 | -0.051 | -0.4336 | No |
| 45 | PSMB5 | PSMB5 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 5 | 9216 | -0.052 | -0.4323 | No |
| 46 | MARCKS | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 9387 | -0.057 | -0.4412 | No |
| 47 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 9553 | -0.061 | -0.4494 | No |
| 48 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 9617 | -0.063 | -0.4497 | No |
| 49 | CFDP1 | CFDP1 Entrez, Source | craniofacial development protein 1 | 9791 | -0.068 | -0.4581 | No |
| 50 | CTBP1 | CTBP1 Entrez, Source | C-terminal binding protein 1 | 9803 | -0.068 | -0.4541 | No |
| 51 | PSMD6 | PSMD6 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | 9834 | -0.069 | -0.4515 | No |
| 52 | HAS2 | HAS2 Entrez, Source | hyaluronan synthase 2 | 9840 | -0.070 | -0.4470 | No |
| 53 | PDCD6IP | PDCD6IP Entrez, Source | programmed cell death 6 interacting protein | 9842 | -0.070 | -0.4422 | No |
| 54 | HNRPL | HNRPL Entrez, Source | heterogeneous nuclear ribonucleoprotein L | 9844 | -0.070 | -0.4375 | No |
| 55 | PRKCDBP | PRKCDBP Entrez, Source | protein kinase C, delta binding protein | 9900 | -0.071 | -0.4366 | No |
| 56 | NR2F2 | NR2F2 Entrez, Source | nuclear receptor subfamily 2, group F, member 2 | 10001 | -0.075 | -0.4389 | No |
| 57 | FSTL1 | FSTL1 Entrez, Source | follistatin-like 1 | 10104 | -0.078 | -0.4411 | No |
| 58 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 10323 | -0.086 | -0.4516 | No |
| 59 | ANP32E | ANP32E Entrez, Source | acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | 10400 | -0.088 | -0.4512 | No |
| 60 | PSIP1 | PSIP1 Entrez, Source | PC4 and SFRS1 interacting protein 1 | 10403 | -0.088 | -0.4452 | No |
| 61 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 10707 | -0.099 | -0.4611 | No |
| 62 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 10778 | -0.102 | -0.4593 | No |
| 63 | EMP3 | EMP3 Entrez, Source | epithelial membrane protein 3 | 10906 | -0.107 | -0.4614 | No |
| 64 | C1R | C1R Entrez, Source | complement component 1, r subcomponent | 10987 | -0.111 | -0.4596 | No |
| 65 | PSMC3 | PSMC3 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 10989 | -0.111 | -0.4520 | No |
| 66 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 11000 | -0.112 | -0.4449 | No |
| 67 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11099 | -0.117 | -0.4441 | No |
| 68 | TIA1 | TIA1 Entrez, Source | TIA1 cytotoxic granule-associated RNA binding protein | 11579 | -0.142 | -0.4703 | Yes |
| 69 | TMEM5 | TMEM5 Entrez, Source | transmembrane protein 5 | 11582 | -0.143 | -0.4605 | Yes |
| 70 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 11589 | -0.143 | -0.4510 | Yes |
| 71 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 11603 | -0.144 | -0.4419 | Yes |
| 72 | SF3A3 | SF3A3 Entrez, Source | splicing factor 3a, subunit 3, 60kDa | 11660 | -0.148 | -0.4358 | Yes |
| 73 | TSEN34 | TSEN34 Entrez, Source | tRNA splicing endonuclease 34 homolog (S. cerevisiae) | 11768 | -0.154 | -0.4331 | Yes |
| 74 | PTOV1 | PTOV1 Entrez, Source | prostate tumor overexpressed gene 1 | 11789 | -0.156 | -0.4237 | Yes |
| 75 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 12013 | -0.174 | -0.4284 | Yes |
| 76 | RBM3 | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 12040 | -0.177 | -0.4180 | Yes |
| 77 | NFIB | NFIB Entrez, Source | nuclear factor I/B | 12064 | -0.179 | -0.4072 | Yes |
| 78 | NUP88 | NUP88 Entrez, Source | nucleoporin 88kDa | 12374 | -0.212 | -0.4158 | Yes |
| 79 | TMPO | TMPO Entrez, Source | thymopoietin | 12402 | -0.215 | -0.4028 | Yes |
| 80 | HAT1 | HAT1 Entrez, Source | histone acetyltransferase 1 | 12419 | -0.217 | -0.3888 | Yes |
| 81 | FMR1 | FMR1 Entrez, Source | fragile X mental retardation 1 | 12443 | -0.220 | -0.3751 | Yes |
| 82 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 12498 | -0.229 | -0.3632 | Yes |
| 83 | CD99 | CD99 Entrez, Source | CD99 molecule | 12537 | -0.234 | -0.3497 | Yes |
| 84 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 12608 | -0.247 | -0.3377 | Yes |
| 85 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 12652 | -0.254 | -0.3232 | Yes |
| 86 | TACC3 | TACC3 Entrez, Source | transforming, acidic coiled-coil containing protein 3 | 12687 | -0.262 | -0.3075 | Yes |
| 87 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 12794 | -0.284 | -0.2956 | Yes |
| 88 | TRIP13 | TRIP13 Entrez, Source | thyroid hormone receptor interactor 13 | 12822 | -0.294 | -0.2771 | Yes |
| 89 | LOXL1 | LOXL1 Entrez, Source | lysyl oxidase-like 1 | 12864 | -0.308 | -0.2586 | Yes |
| 90 | MET | MET Entrez, Source | met proto-oncogene (hepatocyte growth factor receptor) | 12926 | -0.331 | -0.2401 | Yes |
| 91 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12954 | -0.340 | -0.2184 | Yes |
| 92 | IFITM2 | IFITM2 Entrez, Source | interferon induced transmembrane protein 2 (1-8D) | 12966 | -0.345 | -0.1950 | Yes |
| 93 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12999 | -0.364 | -0.1720 | Yes |
| 94 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 13000 | -0.365 | -0.1465 | Yes |
| 95 | CAPG | CAPG Entrez, Source | capping protein (actin filament), gelsolin-like | 13018 | -0.375 | -0.1215 | Yes |
| 96 | HN1 | HN1 Entrez, Source | hematological and neurological expressed 1 | 13064 | -0.406 | -0.0965 | Yes |
| 97 | RCN2 | RCN2 Entrez, Source | reticulocalbin 2, EF-hand calcium binding domain | 13084 | -0.414 | -0.0689 | Yes |
| 98 | AURKB | AURKB Entrez, Source | aurora kinase B | 13112 | -0.438 | -0.0403 | Yes |
| 99 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13288 | -0.823 | 0.0041 | Yes |